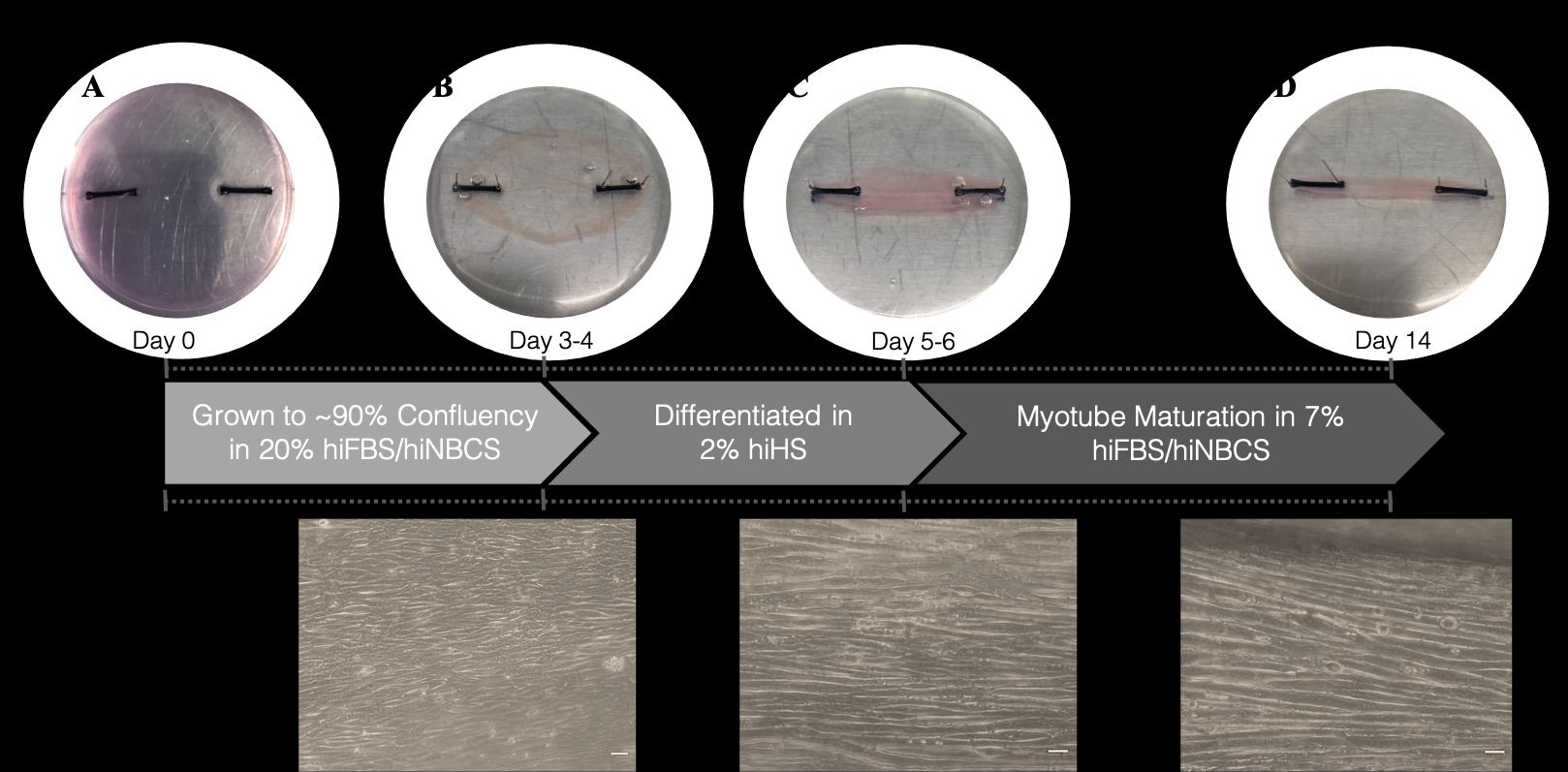
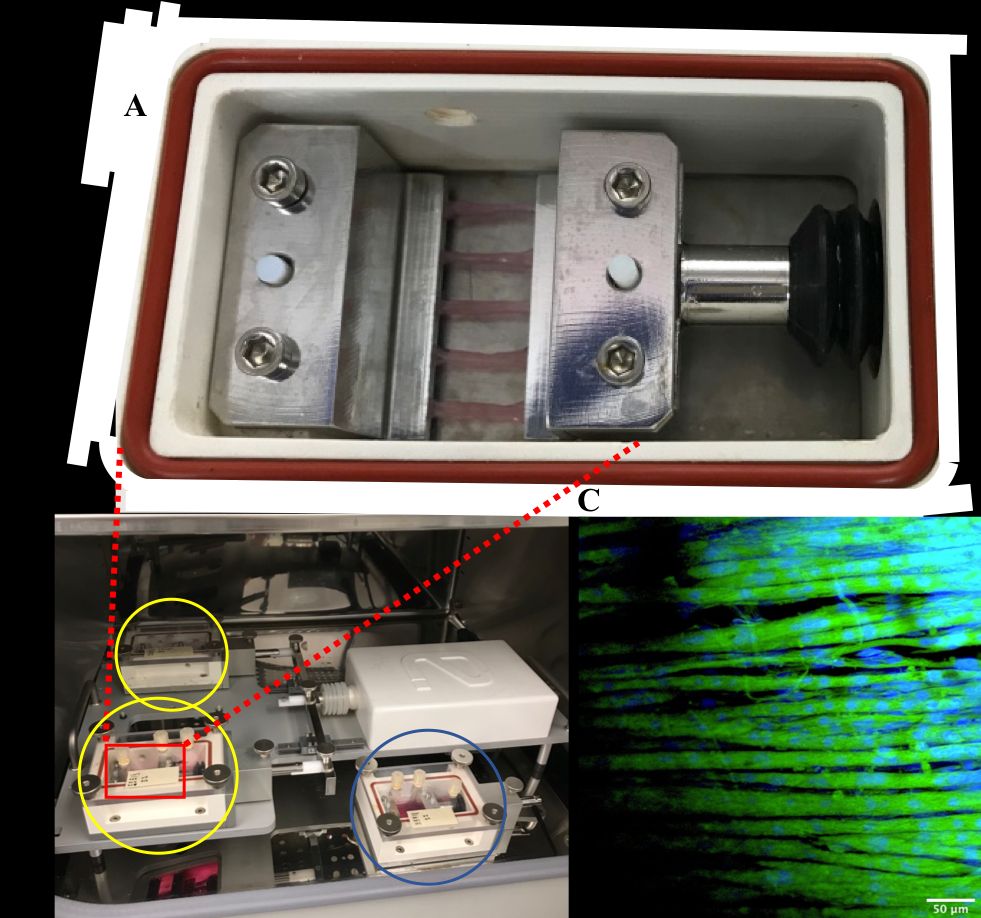
Bioengineered skeletal muscle (SkM) provides an effective model for investigating the molecular mechanisms of mechanical load-induced anabolism and hypertrophy in vitro. However, it is previously unknown whether mechanical loading of bioengineered SkM in vitro adequately recapitulates the molecular responses observed after resistance exercise (RE) in vivo. Furthermore, the epigenetic response to exercise-like stimuli in bioengineered SkM has not previously been determined. To address this, the transcriptional (assessed via RT-PCR) and epigenetic (DNA methylation assessed via targeted next generation bisulfite sequencing, tNGBS) responses were compared after mechanical loading in mouse C2C12 bioengineered SkM (n = 4-5 replicate cultures) in vitro and after RE in vivo (Turner et al., 2021). Specifically, genes known to be upregulated/hypomethylated after RE in humans (n = 8 healthy young men, Seaborne et al., 2018a, 2018b) were analyzed. One‐way ANOVA with post hoc analysis (Tukey HSD) showed that 93% (14/15 genes) of these genes demonstrated similar changes in gene expression post‐loading in the bioengineered muscle when compared to acute RE in humans. Furthermore, similar differences in gene expression of the same genes were observed between loaded bioengineered SkM and after 4 weeks of programmed resistance training (RT) in adult (6 months old) Wistar rats (n = 5 biological replicates, Schmoll et al., 2018). Following unpaired t-test statistical analysis, hypomethylation occurred for only one of the genes analysed (GRIK2) post‐loading in bioengineered SkM. To further validate these findings, DNA methylation and mRNA expression of known hypomethylated and upregulated genes across pooled transcriptomic datasets (110 biopsies, 37 pre and 57 post after outlier removal) after acute RE in humans (Turner et al., 2019) were also analyzed at 0.5-, 3-, and 24-hours post‐loading in bioengineered muscle via a two‐way mixed ANOVA analysis. The largest changes in gene expression occurred at 3-hours, whereby 82% (18/22 genes) and 91% (20/22 genes) of genes responded similarly when compared to human and rodent SkM, respectively. DNA methylation of only a small proportion of genes analyzed (TRAF1, MSN and CTTN) significantly increased post‐loading in bioengineered SkM alone. Overall, mechanical loading of bioengineered SkM in vitro recapitulates the gene expression profile of human and rodent SkM after RE in vivo. Although some genes demonstrated differential DNA methylation post‐loading in bioengineered SkM, such changes across the majority of genes analyzed did not closely mimic the epigenetic response to acute‐RE in humans.
Biomedical Basis of Elite Performance 2022 (University of Nottingham, UK) (2022) Proc Physiol Soc 49, SA12
Research Symposium: Gene expression and DNA methylation after mechanical loading in vitro
Daniel C Turner1, Robert A Seaborne2, Piotr P Gorski1, Mark R Viggars3, Mark Murphy4, Jonathan C Jarvis4, Neil RW Martin5, Claire E Stewart4, Adam P Sharples1
1 Norwegian School of Sport Sciences 2 University of Copenhagen 3 University of Florida 4 Liverpool John Moores University 5 Loughborough University
View other abstracts by:
Where applicable, experiments conform with Society ethical requirements.