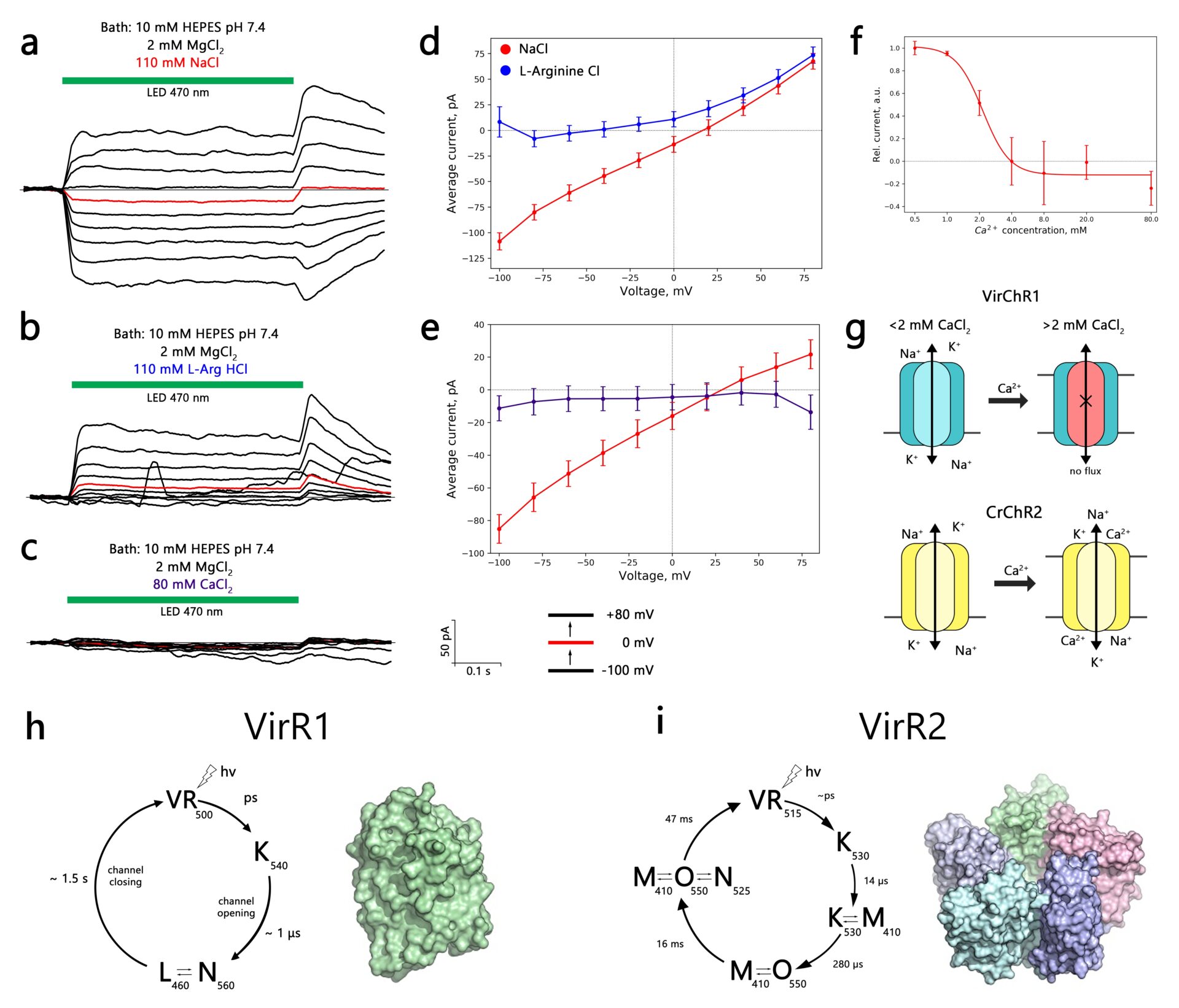
Microbial rhodopsins are a family of light-sensitive membrane proteins that perform various functions upon light illumination. Recently, rhodopsin-like genes were found in genomes of nucleoplasmic large DNA viruses, which opened a discussion about the potential biological implications of the light-sensitive machinery of giant viruses. The viral rhodopsins family comprises two subgroups, namely group 1 (VirR1) and group 2 (VirR2), that differ phylogenetically from non-viral rhodopsins (Yutin and Koonin 2012). We studied members from both groups to shed light on their molecular and cellular functions. Using the patch-clamp method, we tested the electrophysiological properties of VirR-expressing human neuroblastoma cells (SH-SY5Y) in both light and dark conditions. We observed that viral rhodopsins expressed well, but showed strong retention in the cytosol. To address the plasma membrane localization issue, we tested multiple N- and C- terminal modifications of VirR constructs. When we supplemented one of the VirR1 proteins, VirChR1 with p2A self-cleavage peptide prior to fluorescence tag, we were able to get measurable photocurrents. The electrophysiological characterization revealed that VirChR1 is a Na+/K+ selective light-gated ion channel, which can be inhibited by moderate concentrations of Ca2+ ions (~ 2 mM) (Zabelskii et al. 2020). Besides that, we were able to demonstrate that, upon illumination, VirChR1 is able to drive neural firing. Our efforts in the electrophysiological characterization of the VirR2 group did not result in observing any measurable photocurrents.
In order to gain more insight into the molecular function of viral rhodopsins, we expressed, purified, and characterized OLPVR1 and VirChR1 rhodopsins from the VirR1 group, and OLPVRII rhodopsin from the VirR2 group. Upon light illumination, OLPVRII rhodopsin undergoes a photocycle with 70 ms duration, which indicates pump-like behavior, whereas, both OLPVR1 and VirChR1 have channel-like photocycle with a duration of around several seconds. OLPVR1 and OLPVRII proteins were crystallized using in meso crystallization method and have yielded high-resolution structures of 1.4 Å and 1.9 Å respectively. Due to the conservativity of viral rhodopsins, the structures provide structural insight into their potential function. OLPVRII forms a pentamer, with a symmetrical, bottle-like central channel with a narrow vestibule in the cytoplasmic part covered by a ring of 5 arginines, whereas 5 phenylalanines form a hydrophobic barrier in its exit (Bratanov et al. 2019). The putative central channel is blocked by a hydrophobic tail of lipid from the crystallization matrix that potentially prevents the channel-like function of the protein. OLPVR1 crystallizes as a monomer and shares many structural features with well-studied channelrhodopsin 2 from Chlamydomonas reinhardtii (Volkov et al. 2017). OLPVR1 has three consecutive constriction sites that facilitate ion transport upon photon absorption. The OLPVR1 protomer has short extracellular loops, which sharply differentiates it from other channelrhodopsins that typically have large N- and C-terminal domains (Ernst et al. 2014). We are currently looking for ways to improve the plasma membrane localization of viral rhodopsins that can help to understand the function of OLPVRII and help viral rhodopsins to find their niche in optogenetics applications.