Introduction: The structure and function of cardiomyocytes (CM) are tightly interlinked. However, ultrastructural dynamics of CM during the cardiac action potential, including mechanical organelle deformation, are poorly understood. Dynamics of contracting CM are conventionally resolved using light microscopy, a modality with orders of magnitudes lower spatial resolution than electron-based imaging.
Aims: The aim of this study is to develop and apply a methodology for the acquisition and analysis of ultrastructural dynamics of CM under control conditions and following microtubule (de-)stabilisation. Through this study, we aim to gain a better understanding of the ultrastructural dynamics of CM during cardiac contraction, which may have important implications for our understanding of cardiac function and mechanosensitivity.
Methods: Here, we use action potential-synchronised high-pressure freezing to assess the ultrastructural dynamics during CM contraction with dual-axis electron tomography, resulting in a spatial resolution of (1.2 nm)3 and millisecond temporal resolution. CM were isolated from precision-cut left-ventricular New Zealand white rabbit tissue slices (N=8 animals). We used pharmacological interventions (paclitaxel and colchicine) to stabilise and destabilise microtubules. CM were high-pressure-frozen at time intervals corresponding to rest and peak contraction, freeze-substituted, heavy metal-stained, resin-embedded, and cut into 300 nm sections. Then, CM fragments were imaged using electron tomography on a 300 kV transmission electron microscope. The resulting images were reconstructed and segmented utilising fully convolutional neural networks into 3D organelle models. We developed custom software ('SegmentPuzzler') to proofread and correct automatic segmentations. We developed a portable, interactive browser-based visualization tool to foster a deeper comprehension of the otherwise unwieldy (TB-sized) image data and reconstructions. Statistical significance was assessed using the Kruskal-Wallis test and Dunn’s posthoc test. All investigations reported in this article conformed to the German (TierSchG and TierSchVersV) animal welfare laws, compatible with the guidelines stated in Directive 2010/63/EU of the European Parliament on the protection of animals used for scientific purposes, and they were approved by the local Institutional Animal Care and Use Committees in Germany (Regierungspräsidium Freiburg, X-16/10R).
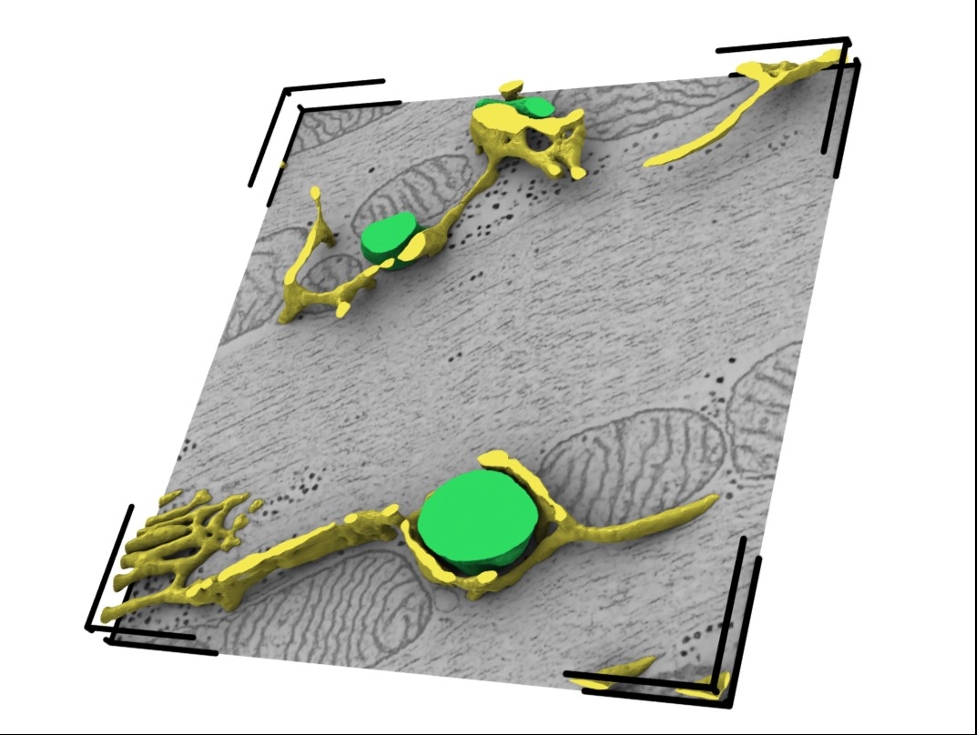
Results: Using this workflow, we generated and visualised 353 3D reconstructions of CM organelles, including the sarcoplasmic reticulum and the transverse-axial tubular system (Figure 1). We analysed dyad dimensions, coupling distances, and deformation of the t-tubular geometry. In control conditions, we observed a t-tubular squeezing effect (p<0.0001), which could not be detected after the acute (de-)stabilization of microtubules (p>0.05). The presence or absence of microtubules had no significant acute effect on dyad proximity (p>0.05).
Conclusion: Our proof-of-principle study resolves the structural dynamics of CM in a nanoscopic, 3D, and millisecond-accurate manner. Precisely understanding the ultrastructure and its modulation, ultimately in human CM under both physiological and pathophysiological conditions, is expected to advance our current understanding of the ultrastructural foundations of cardiac diseases, and their diagnosis and treatment.
Figure 1: Time-resolved 3D organelle model of the sarcoplasmic reticulum (yellow) and transverse-axial tubular system (green). Cardiomyocytes in this reconstruction were high-pressure-frozen in a relaxed state (0 ms offset to the action potential initiation). The reconstruction has a dimension of 3.14 µm x 3.14 µm x 180 nm and a voxel size of (1.2 nm)3.