Physiology News Magazine
Polygenic scores and precision genetics
Polygenic scores: what they are, how they are calculated and how they can facilitate clinical outcomes
Features
Polygenic scores and precision genetics
Polygenic scores: what they are, how they are calculated and how they can facilitate clinical outcomes
Features

Agnieszka Gidziela
School of Biological and Behavioural Sciences, Queen Mary University of London
What is a polygenic score?
A polygenic score, or polygenic index, is a mathematical tool that scientists use to estimate the combined effects of many different genes on a specific trait or characteristic. Polygenic scores were originally utilised in plant and animal breeding and transitioned to human genetics in 2007 when researchers proposed using these scores to identify individuals at an elevated risk of disease (Wray et al., 2007). The practical application took shape in 2009 through a study on schizophrenia that was not only the first one to apply polygenic scores, but also introduced the term “polygenic score” (Purcell et al., 2009).
Complex pathological conditions in humans tend to be associated with not one, but many genetic variants, each playing a small yet different role in increasing or decreasing risk. Polygenic scores provide a way of bringing together genetic risk scattered across the entire genome. They take findings from large gene-discovery studies and aggregate information from up to a few million genetic variants into a single score that provides a personalised index, although partial, of genetic influence on a trait. It is possible to calculate a polygenic score for any trait for which a gene-discovery study is available—physical, health-related, or psychological. A way to think about polygenic scores might be as a graded measure of risk calculated directly from DNA; not unlike a credit score provides a partial summary of your financial risk, a polygenic score provides a partial index of your genetic risk for a particular disorder or trait. Polygenic scores are a valuable tool in precision medicine as they not only help to tailor screening, prevention, and treatment strategies based on an individual’s genetic risk profile but also provide a more nuanced understanding of risk and outcomes, allowing for more personalised healthcare choices, including decisions about risk management strategies.
How are polygenic scores calculated?
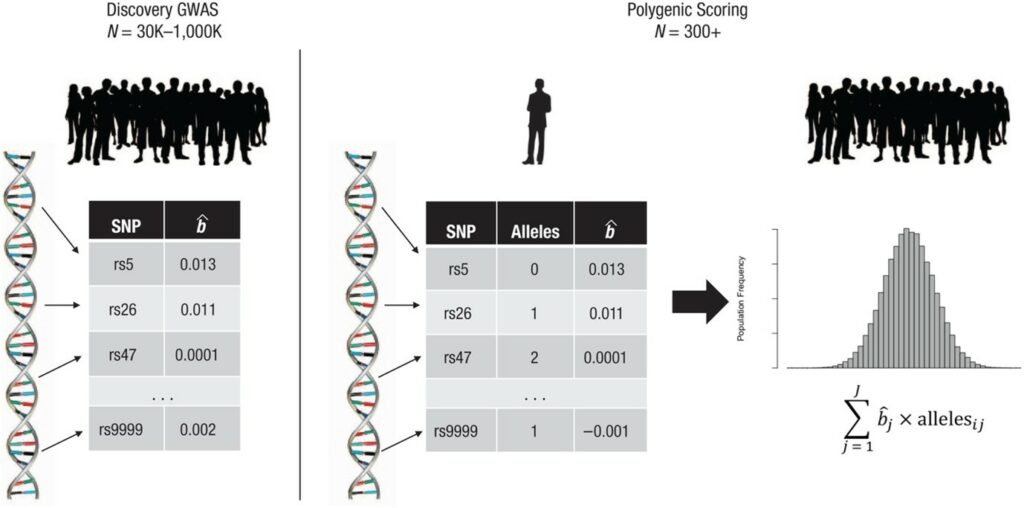
Polygenic scores are calculated using data from genome-wide association studies (GWAS). These are large gene-discovery studies that scan the genome (i.e. decoded DNA sequence) of several hundred thousand or even millions of people to identify genetic variants, namely the single nucleotide polymorphisms (SNPs), that are associated with a specific disease or physical or behavioural characteristic. When adequately powered, GWAS can reliably identify genetic variants that are associated with a given trait as well as how strongly each variant is associated with differences between people in that trait (i.e. the effect size of their association). The effect size can be positive, therefore conferring an increased risk of developing a disorder or trait, or negative therefore lowering the risk. To calculate a polygenic score, researchers combine all the genetic variants that are associated with a given trait or disorder, each weighted by its effect size. The process of creating polygenic scores using data from GWAS is illustrated in Fig.1, derived from Belsky and Harden (2019), where the participant’s SNPs are given weights that signify the direction and strength of the link to a specific trait. Typically, these weights are derived from effect sizes, denoted as bˆj, estimated in a GWAS that did not involve the research participant. Following this, the count of alleles associated with the phenotype at each SNP (j) is estimated, and this count is multiplied by the corresponding weight, bˆj. Ultimately, the weighted counts are added together across all SNPs to determine the participant’s polygenic score. The resulting distribution of polygenic scores among participants follows a normal distribution, corresponding to the distribution of symptoms of a particular disease (Fig.2).
How accurate are polygenic scores?
While lifestyle factors are important contributors to many health-related outcomes, genetics also play a significant role in determining an individual’s susceptibility, hence polygenic scores offer insights beyond lifestyle factors. Genetic risk assessments can help to identify individuals at risk for certain conditions early in life, sometimes before lifestyle-related issues become apparent. This early identification allows for proactive and preventive measures. In addition, certain health conditions have a strong genetic component that may contribute to risk even in the absence of risky lifestyle behaviours. Polygenic scores help identify individuals who may be at elevated risk due to their genetic makeup. Some polygenic scores are more accurate than others in terms of predicting real-life outcomes. For cardiovascular disease, for example, studies have shown that a high polygenic score can be as effective at predicting the risk of heart problems as other lifestyle risk factors that are usually considered by medical practitioners, for example, smoking and body mass index (Inouye et al., 2018).
Another condition for which polygenic scores could be of great utility is diabetes. Using polygenic scores, it might be possible to diagnose young adults with diabetes who might need insulin treatment (Padilla- Martínez et al., 2020) and to differentiate between different types of diabetes (e.g. type 1 versus type 2) (Oram et al., 2016). With such a high level of accuracy, greater precision in diagnosis, facilitated by polygenic scores, could result in lifestyle changes or targeted use of medication to effectively treat, or even slow down the progression of the condition before the symptoms have manifested.
Polygenic scores have been proven to be valuable not only in predicting physiological outcomes but also in shedding light on behavioural traits. However, compared to other physical traits or medical conditions, like diabetes or coronary artery disease, the prediction of behaviour and emotional problems remains relatively small, predicting less than 6% of differences between children and adolescents (Gidziela et al., 2022). Although the current level of accuracy means that polygenic scores are not yet useful in correctly identifying children at risk of behaviour problems, they are currently a valuable tool in behavioural research. For example, researchers are interested in exploring whether combining polygenic scores with other environmental risk factors might lead to greater prediction accuracy.
It is important to note that while polygenic scores offer insights into potential relationships between genetics and health outcomes, they’re not perfect predictors. Many factors shape our behaviour and health, including life experiences and environment. Nevertheless, polygenic scores provide a valuable tool for researchers to better understand the complex interplay between genetics, health, and behaviour, helping us uncover more about the factors that contribute to differences in traits and diseases. In essence, polygenic scores provide insightful clues, but they’re not crystal-clear indicators just yet. We are on a journey of refining and improving them to enhance their accuracy for practical applications.
When talking about prediction accuracy of polygenic scores it cannot be separated from the extent to which a trait is shaped by genetics, known as its heritability. Essentially, heritability measures how much of a particular trait’s variation, i.e. individual differences in that trait, is attributable to genetic variation. It is crucial to understand that when dealing with complex traits that are only partly influenced by genetics, such as physiological and behavioural traits, the predictive models based solely on genetic information tend to be less accurate (Plomin and Von Stumm, 2021). Most complex traits are influenced by both genetics and the environment, leading to a complex interplay.
A fascinating aspect of polygenic scores is that they’re like a static snapshot of the genome; they provide a picture that doesn’t change over time because the structure of the DNA does not change. This means they can be measured at any time, unlike some other biological markers that might change, for example, DNA methylation, a marker of gene expression. Nevertheless, this does not mean that polygenic scores are free from environmental effects. In fact, polygenic scores are constructed based on phenotypes that always vary as a function of both genetics and the environment. For example, scientists have observed that the polygenic prediction of educational achievement tends to strengthen as individuals progress through their educational journey, that is likely to reflect a correlation between genes and environments (Allegrini et al., 2019). As children mature, they actively shape their experiences and environments, influenced in part by their genetic propensities, and these experiences, in turn, impact their academic progress.

What are the limitations of polygenic scores?
Polygenic scores, while a valuable tool in genetics and genomics, do have several limitations.
The precision of polygenic scores hinges on the quality and diversity of the genetic data employed in their calculation. In cases where the underlying datasets show biases or a lack of diversity, the predictive accuracy of these scores can be compromised. Such biases can happen when some populations are much more common in the data than others. When this occurs, it can lead to inaccurate risk assessments and predictions for underrepresented or marginalised groups. This limitation underscores the importance of diverse and representative genetic datasets to ensure reliable application of polygenic scores across different populations (Martin et al., 2018).
Furthermore, complex traits often result from an intricate interplay between genetics and environmental exposures. Polygenic scores solely capture the genetic component of these traits and diseases, disregarding the substantial influence of environmental variables that are also relevant for disease development. While genes play a role, they are just a piece of the puzzle and miss out on these external aspects. Therefore, polygenic scores cannot be used to make definitive predictions about an individual’s risk of developing a disease (Wray et al., 2021).
Another limitation includes incomplete understanding of the genetic underpinnings of various traits and diseases. Polygenic scores rely on the knowledge of known genetic variants associated with a particular trait or condition. However, the genetic landscape is far from fully explored, and polygenic scores do not encompass all relevant genetic factors. The present state of genetic knowledge underscores the need for cautious interpretation and application of polygenic scores (Wray et al., 2021).
In addition, use of polygenic scores in healthcare and decision-making contexts raises ethical concerns related to privacy, discrimination, and the potential for stigmatisation. Ensuring responsible and equitable use of polygenic scores is an ongoing challenge (Lewis and Green, 2021; Wray et al., 2021).
There is also the question of whether whole-genome-sequencing (WGS) is required to generate polygenic scores for its use in routine medical care. WGS involves sequencing of the entire genome. While WGS provides comprehensive genetic information, it is costly and may not be necessary for generating polygenic scores. Despite the present cost of WGS being a few thousand dollars, the expenses are rapidly decreasing and are anticipated to dip below $1000 (Plomin and Simpson, 2013).
The utility of polygenic scores in routine medical care depends on the specific application. Polygenic scores may be more relevant for complex traits influenced by multiple genetic factors rather than single-gene disorders. Before polygenic scores become routine in medical care, regulatory bodies and healthcare providers need to establish guidelines for their use. This includes considerations of accuracy, clinical validity, and ethical considerations.
What are the potential applications of polygenic scores?
Polygenic scores have a number of potential applications. They can be used to identify individuals who are at an increased risk of developing a disease. This information can be used to develop preventive measures or to provide early diagnosis and treatment. Polygenic scores are a promising tool for understanding and preventing disease. However, it is important to remember that they have limitations and should not be used as the sole basis for making medical decisions.
We will discuss specific potential applications of polygenic scores for physiologists, focusing on breast cancer, cardiovascular health and disease risk, pharmacogenetics, metabolic health, precision medicine and personalised interventions.
Polygenic scores play a significant role in breast cancer research and clinical practice. They help identify women at higher genetic risk for breast cancer, enabling tailored population screening and risk assessment for those without known genetic mutations (Yanes et al., 2020). Polygenic scores also enhance risk estimation for individuals with genetic mutations, influencing decisions about preventive measures, such as prophylactic mastectomy (Yanes et al., 2020). Additionally, polygenic scores distinguish between breast cancer subtypes and predict diagnostic outcomes, assisting women with treatment decisions based on their history and prognosis (Yanes et al., 2020).
Beyond breast cancer, research has shown that polygenic scores can provide insights into an individual’s genetic predisposition to cardiovascular conditions like heart disease, stroke, and high blood pressure (Abraham et al., 2021). By analysing specific genetic variations associated with these conditions, cardiologists can better understand why some people are more vulnerable and develop strategies for early detection and tailored prevention plans.
Polygenic scores can also be used to study drug response and pharmacogenetics– a study of how genes influence how our bodies respond to medications. Polygenic scores can help predict how a person might metabolise and respond to specific drugs (Fabbri and Serretti, 2020). This knowledge helps avoid adverse reactions and optimise treatment outcomes.
Understanding how genetics influence metabolic health is another area of interest for physiologists, because polygenic scores offer insights into an individual’s risk of developing diabetes. By identifying genetic variants linked to glucose regulation and insulin sensitivity, researchers can gain a better understanding of why some individuals are more prone to metabolic disorders (Padilla-Martínez et al., 2020).
Polygenic scores have the potential to contribute to the field of precision medicine. By combining genetic information with other health data, physiologists can develop personalised intervention plans that consider an individual’s genetic predispositions (Wray et al., 2021). For instance, they can identify individuals who are at higher risk for certain conditions and recommend targeted interventions for prevention.
The future of polygenic scores to guide more personalised healthcare
It is nonetheless essential to acknowledge that polygenic scores are not flawless. They cannot definitively predict the occurrence or absence of a disease in an individual. In essence, polygenic scores offer physiologists a powerful tool to guide the development of tailored interventions, advance our understanding of human physiology, and pave the way for more personalised approaches to healthcare. However, before this is implemented in routine healthcare, the field has to overcome limitations such as representative bias (mostly European populations) in genomic datasets, understanding how to account for environmental and lifestyle factors in prediction scores, creating and/or selecting statistical models that best reflect specific patterns of a disease and tackling current ethical concerns of using unvalidated polygenic scoring processes in commercial settings, to name a few.
References
Abraham G et al. (2021). Risk prediction using polygenic risk scores for prevention of stroke and other cardiovascular diseases. Stroke 52(9), 2983–2991.
Allegrini AG et al. (2019). Genomic prediction of cognitive traits in childhood and adolescence. Molecular Psychiatry 24(6), 819–827.
Belsky DW, Harden KP (2019). Phenotypic annotation: using polygenic scores to translate discoveries from genome-wide association studies from the top down. Current Directions in Psychological Science 28(1), 82–90.
Fabbri C, Serretti A (2020). Clinical application of antidepressant pharmacogenetics: considerations for the design of future studies. Neuroscience Letters 726, 133651.
Gidziela A et al. (2022). Using DNA to predict behaviour problems from preschool to adulthood. Journal of Child Psychology and Psychiatry 63(7), 781–792.
Inouye M et al. (2018). Genomic risk prediction of coronary artery disease in 480,000 adults: implications for primary prevention. Journal of the American College of Cardiology 72(16), 1883–1893.
Lewis AC, Green RC (2021). Polygenic risk scores in the clinic: new perspectives needed on familiar ethical issues. Genome Medicine 13, 1–10.
Oram RA et al. (2016). A type 1 diabetes genetic risk score can aid discrimination between type 1 and type 2 diabetes in young adults. Diabetes Care 39(3), 337–344.
Padilla-Martínez F et al. (2020). Systematic review of polygenic risk scores for type 1 and type 2 diabetes. International Journal of Molecular Sciences 21(5), 1703.
Plomin R, Simpson MA. (2013). The future of genomics for developmentalists. Development and Psychopathology (in press).
Plomin R, Von Stumm S (2021). Polygenic scores: prediction versus explanation. Molecular Psychiatry 27, 49–52.
Purcell SM et al. (2009). Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature, 460(7256), 748–752.
Wray NR et al. (2021). From basic science to clinical application of polygenic risk scores: a primer. JAMA Psychiatry 78(1), 101–109.
Wray NR et al. (2007). Prediction of individual genetic risk to disease from genome-wide association studies. Genome Research 17(10), 1520–1528.
Yanes T et al. (2020). The emerging field of polygenic risk scores and perspective for use in clinical care. Human Molecular Genetics 29(R2), R165–R176.
