Introduction
The dynamics of human voluntary muscle contraction are commonly investigated with a mathematical Hill-type model that transforms a single bipolar EMG signal into whole muscle force. However, this single-input single-output approach limits the comprehensive description of muscle internal dynamics, like the recruitment and discharge dynamics of the muscle’s individual motor units (MUs), as reviewed [1]. Here, we propose a novel and validated mathematical model [2] that takes as input a vector of experimental motoneuron (MN) spike trains to control, in MN-driven simulation of human voluntary muscle contractions, the force-generating dynamics of a population of individual MUs.
Methods
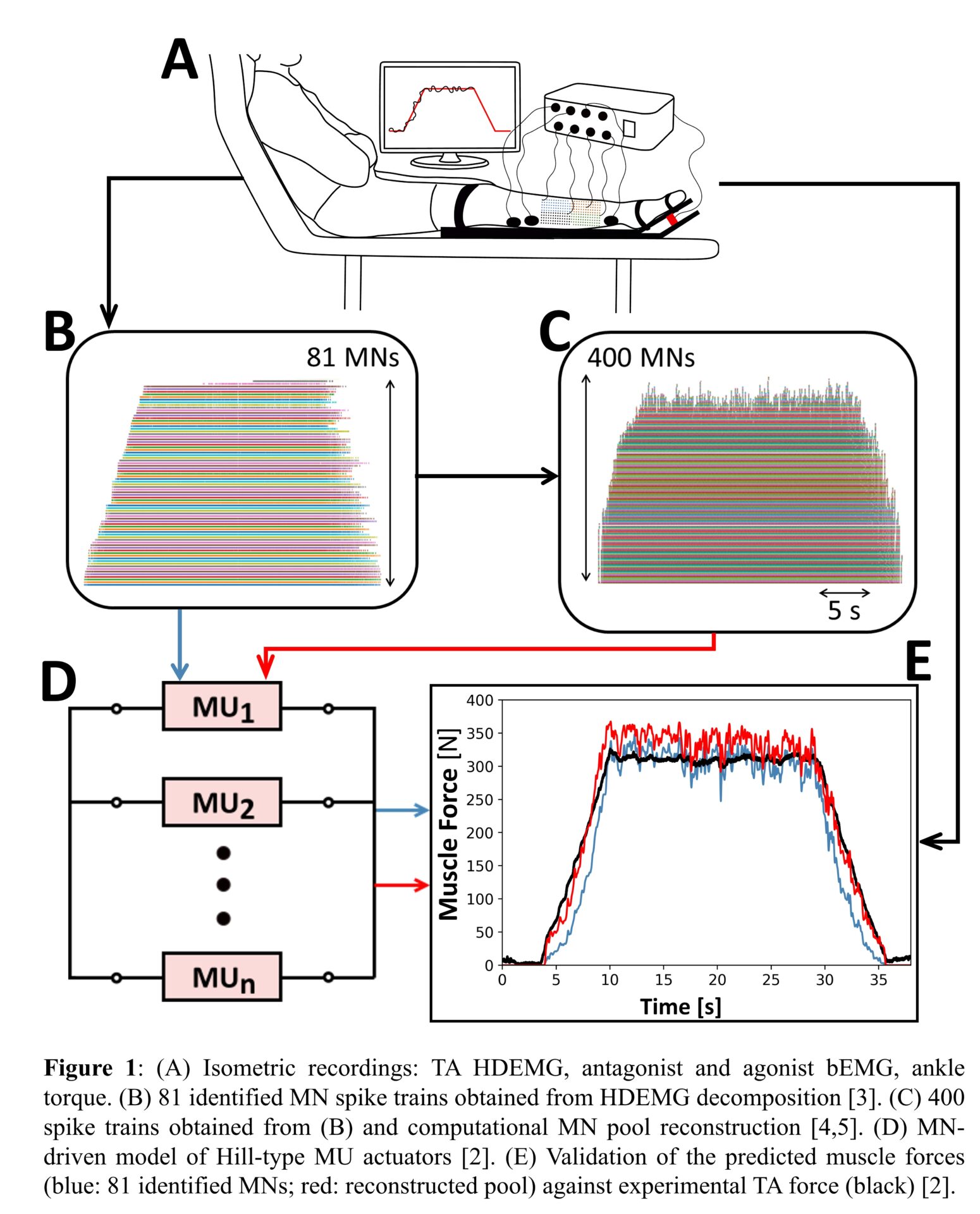
The model is built as a population of Hill-type actuators, each of which describes the force-generating dynamics of an individual MU. This approach is MN-driven: the pool of Hill-type MUs transforms a vector of binary MN spike trains into a vector of continuous MU forces, that sum into the predicted whole muscle force (Figure 1). By using a musculoskeletal model derived from segmented medical images, carefully-selected experimental biophysical parameters from previous literature, and advanced modelling techniques, the MU forces are derived from the input spike trains with subject-muscle-specific models of the dynamics of free calcium concentration, concentration of calcium-troponin molecules, MU active state, and other MU mechanical properties. Note that no parameter calibration was performed.
The input vector of MN spike trains was derived from blind source separation of HDEMG signals (Figure 1A, B) acquired during an isometric trapezoidal contraction up to 30% of the maximum force of the tibialis anterior (TA) muscle of a healthy subject (male, 26 years old, 170 cm, 52 kg) [3]. The sensitivity of the model performance to the experimental motoneuronal data was assessed by varying the size and the density of the grids of EMG electrodes [3] and by reconstructing the discharge activity of the complete MU pool with computational methods (Figure 1C) [4,5].
To evaluate the prediction accuracy of the multi-MU model, eight validation metrics, including the normalized root-mean-squared error (NRMSE) and coefficient of determination (r2), were computed between the predicted and measured muscle forces.
Results and Discussion
The best prediction accuracy was obtained when the input vectors of MN spike trains were representative of the full spectrum of discharging MUs and accurately approximated the true neural drive to muscle. Yet, few to no early-recruited MUs are usually identified with HDEMG decomposition. This experimental limitation was addressed by using large and dense grids of EMG electrodes (10×3.6cm, 256 electrodes) (Figure 1B) [3] or by computationally reconstructing the whole MU population (Figure 1C) [4,5]. In the best conditions, the predicted and measured TA forces (Figure 1E) were in remarkably good agreement (r2=0.99, NRMSE=6%).
Conclusions
This state-of-the-art neuromuscular model samples the predicted whole muscle force into individual MU dynamics controlled by specific experimental spike trains. As opposed to single-actuator bEMG-driven Hill-type models, this comprehensive description of the MU recruitment and discharge activity in human voluntary contraction finds dedicated applications in the control of neuroprosthetics, the investigation of neural synergies, and volumetric muscle modelling.