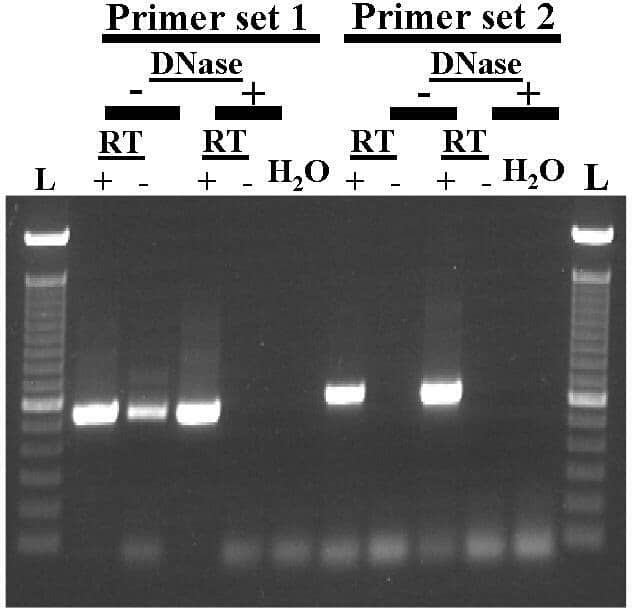
β–actin is often favoured as a reference gene in transcript expression studies. However, when using human samples, e.g. placental or uterine biopsies, the small tissue mass tends to give low total RNA yields that are further reduced by DNaseI treatment to remove genomic DNA. A further problem if not DNaseI–treating such samples is the possible co–amplification of processed pseudogenes from genomic DNA. Therefore, in RT–PCR analysis of small human uterine smooth muscle samples, we have compared the efficacy of traditional exon–intron spanning primers for β–actin (Primer set 1: forward 5′–ggg acc tga ctg act acc tc–3′ and reverse 5′–act cgt cat act cct gct tg–3′; Dye et al. 2004), with and without DNaseI treatment, to primers suggested to only recognise reverse transcribed β–actin and not pseudogenes (Primer set 2: forward 5′–cct cgc ctt tgc cga tcc–3′ and reverse 5′–gga tct tca tga ggt agt cag tc–3′; Raff et al. 1997). Total RNA was extracted from human myometrial biopsies, obtained at Caesarean section following written informed consent, using Trizol. RNA samples (n=3) were either digested with DNaseI or left untreated. cDNA synthesis was performed in the presence (RT+) and absence (RT−) of reverse transcriptase and PCR performed using both sets of β–actin primers. The exon–intron spanning primers of Dye et al. (2004) produced signals of similar size in both the (RT+) and (RT−) lanes in DNaseI–untreated samples indicative of the amplification of pseudogenes (Fig 1). DNaseI–treated samples only produced signals in (RT+) lanes indicating successful removal of genomic DNA from the total RNA. In contrast, the β––actin primers of Raff et al. (1997) only produced signals in (RT+) lanes even in those samples contaminated with genomic DNA (Fig. 1). This confirms the usefulness of these primers in only detecting reverse transcribed β–actin product, and not any coamplified pseudogene product, from human uteri. This is of particular use when analysing β–actin RNA from small amounts of precious starting material that one doesnt wish to exhaust with additional purification procedures. However, in such DNaseI–untreated human samples it will remain important, for detecting other genes of interest containing more than one exon, to design primers that span exon–intron junctions. Fig. 1. Comparison of RT–PCR with 2 β–actin primer sets for human myometrial RNA samples ±DNaseI treatment. RT =reverse transcriptase, L = DNA 100–2000 bp Ladders. –ve control = H20.
King's College London (2005) J Physiol 565P, PC179
Communications: Reliable Detection of β–Actin Transcript from Human RNA Samples Contaminated with Genomic DNA
Taggart, Michael John; Halliday, Deborah ; Lacey, Helen ;
1. Cardiovascular Research, University of Manchester, Manchester, United Kingdom. 2. Maternal and Fetal Health Research Centre, University of Manchester, Manchester, United Kingdom. 3. Academic Unit of Child Health, University of Manchester, Manchester, United Kingdom.
View other abstracts by:
Where applicable, experiments conform with Society ethical requirements.