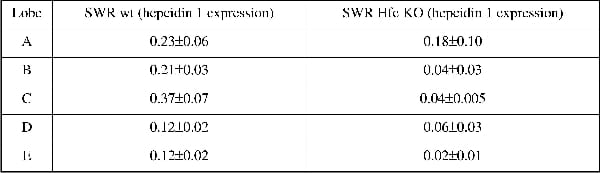
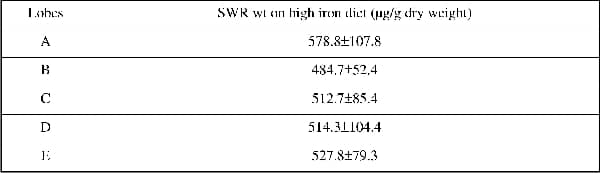
Hereditary haemochromatosis is an autosomal recessive disorder of iron (Fe) metabolism and a mutation in the Hfe gene is the most common cause of the disease. Haemochromatosis is characterised by defective regulation of dietary iron absorption that leads to excessive Fe accumulation in various organs including the liver, pancreas, heart, joints, and pituitary gland leading to hepatic cancer, liver failure, diabetes, and heart disease. Recent findings suggest hepcidin, a hormone produced by the liver, is a negative regulator of Fe absorption and is itself regulated by iron, anaemia, hypoxia and inflammation. In the case of haemochromatosis expression of hepcidin is inappropriate for the levels of liver Fe and this leads to increased Fe uptake despite liver iron being elevated. These observations are based on measurement of iron and hepcidin expression in biopsies or a part of a lobe; however, there is no information on Fe levels or hepcidin expression in relation to different lobes in the liver. The aim of this study was to investigate the level of Fe in each lobe of the liver and measure the hepcidin gene expression in SWR-wild type (wt) and SWR-Hfe knockout mice. Both wt and Hfe knockout mice were studied at the age of 12 weeks. The livers were divided into five lobes and named A to E, Fe quantification was done by the Torrance and Bothwell method and hepcidin gene expression was measured by RT-PCR. We found that in SWR wt mice liver Fe in all the lobes ranged from 70 to 90 µg/g dry weight (79.8±5.1 µg/g dry weight). In SWR Hfe knockout mice, liver Fe levels were higher as expected and in lobes B to E Fe levels ranged from 350 to 500 µg/g dry weight (438±15.6 µg/g dry weight). Interestingly, lobe A had a much higher level of Fe than any of the other lobes (821.6±102.8 µg/g dry weight, n=8). When hepcidin 1 gene expression levels were measured, there was no difference between the different lobes in the SWR wt mice (Table 1), but in the SWR Hfe knockout mice, although the hepcidin levels were lower than the wt, there were differences between the lobes, lobe A being the one with the highest level of hepcidin expression. When SWR wt mice were fed a high Fe diet, although there was a great accumulation of Fe in the liver, no differences were seen between the lobes (Table 2). In conclusion, the Hfe gene seems to cause a preferential accumulation of Fe in the specific lobes of the liver. Furthermore hepcidin expression was highest in the lobe with the highest Fe levels. This suggests that liver Fe levels regulate hepcidin but this regulation is perturbed by Hfe. This finding is also relevant to the diagnosis of haemochromatosis as the common practice is to perform biopsies of a particular region of the liver and this might not correspond to the actual Fe accumulation status of the liver.
University College London 2006 (2006) Proc Physiol Soc 3, PC186
Poster Communications: Genetic iron overload versus dietary iron overload in SWR mice
Monica Mascarenhas1, Kaila S Srai1, Robert Simpson2
1. Biochemistry & Molecular Biology, Epithelial Transport and Cell Biology Group (ETCBG), University College London (Hampstead Campus), London, United Kingdom. 2. Life Sciences, King's College London, London, United Kingdom.
View other abstracts by:
Table 1. Hepcidin 1 mRNA content relative to actin in the different liver lobes of SWR wt (n=5) and Hfe knockout (n=5) fed on a normal diet
Table 2. Liver irons (µg/g dry weight) of the different lobes in SWR wt fed an high iron diet (n=6)
Where applicable, experiments conform with Society ethical requirements.