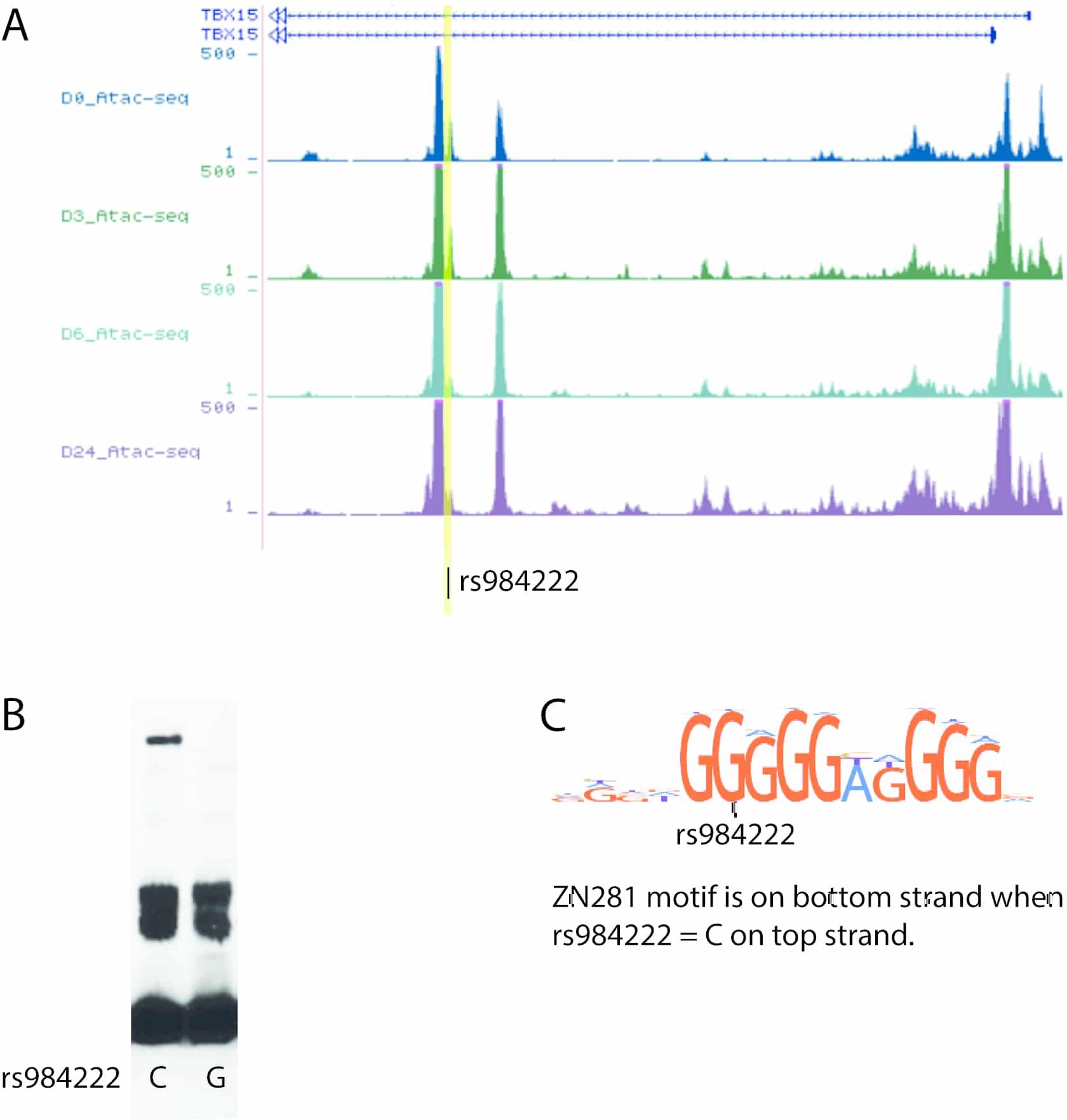
While overweight and obesity carry increased risk of metabolic disease, the distribution of excess body fat, measured indirectly by waist-hip ratio and independent of body mass index, is a predictor of metabolic disease risk. Genome-wide association studies (GWAS) have so far identified 346 loci associated with waist-hip ratio, including the WARS2-TBX15 locus (1, 2). We aim to determine the mechanism(s) by which this locus may alter adipose tissue development and function in different fat depots. The WARS2-TBX15 locus has been further fine-mapped in a study combining populations of European and African descent to an approximately 1.5 kb region within the first intron of TBX15, which includes the lead single nucleotide polymorphism (SNP) from the initial publication of this locus, rs984222 (3). We show that this SNP overlays a peak in an assay for transposase-accessible chromatin (ATAC)-seq experiment in a human pre-adipocyte cell line (hWAT) throughout differentiation and in mature cells (figure 1A); indicating that this region is accessible to transcription factor binding. We demonstrate altered binding of nuclear protein from differentiating (day 4) hWAT cells to DNA oligos containing either version of rs984222 with electrophoretic mobility shift assays (figure 1B; 4 independent replicate experiments, band is lost when SNP=G), corresponding to predicted loss of a ZN281 binding motif (figure 1C). The human 1.5kb fine-mapped region corresponds to an 841bp region in intron 1 of Tbx15 in the mouse and we demonstrate enhancer activity of this region in a mouse pre-adipocyte cell line (3T3L1) throughout differentiation using luciferase enhancer assays. N=8, fold change luciferase activity vs scrambled control; differentiation day -2, 1.97±0.18 P<0.0001; day 0, 1.76±0.17 P=0015; day 1, 1.58±0.10 P=0.014; day 4, 1.93±0.28 P=0.0001 (arbitrary units, mean±SEM, unpaired t-test, representative data of three experiments). These data suggest a role of this fine-mapped region in transcriptional regulation, and supports the use of mouse models for investigating human GWAS identified loci. We have produced a CRISPR mouse model lacking this enhancer region and will present preliminary metabolic phenotyping data from this line; including body weight, fat mass and lean mass (EchoMRI), in comparison to a spontaneously occurring Tbx15-/- mouse where the deletion begins in intron 1 downstream of the 841bp fine-mapped region (4, 5). This is along with planned RNA-seq experiments on tissue from different fat depots in these mice which will help inform the regulatory network of TBX15 and that potentially regulated by this putative enhancer region in intron 1 of the gene.
Physiology 2019 (Aberdeen, UK) (2019) Proc Physiol Soc 43, PC157
Poster Communications: Investigating enhancer activity of a finemapped WARS2-TBX15 GWAS locus associated with waist-hip ratio
R. Dumbell1, M. Muso1, L. Zolkiewski1, S. Laber1,2,3, L. Bentley1, R. Cox1
1. Mammalian Genetic Unit, MRC Harwell Institute, Didcot, Oxon, United Kingdom. 2. The Broad Institute, Cambridge, Massachusetts, United States. 3. The Big Data Institute, University of Oxford, Oxford, United Kingdom.
View other abstracts by:
<p style="margin: 0px 0px 10.66px; text-align: justify;"><font color="#000000" face="Calibri" size="3">A: ATAC-seq peaks surround rs984222 in intron 1 of <i style="mso-bidi-font-style:normal">TBX15 at in a human preadipocyte cell line (hWAT) on differentiation day (D) 0, 3, 6, and 24 (mature adipocytes). B: Representative electrophoretic-mobility shift assay of rs984222 biotin labelled oligo with C and G version of SNP and day 4 differentiated hWAT cell nuclear protein. C: Predicted ZN281 motif on bottom strand is not predicted when rs984222=G.</font></p>
Where applicable, experiments conform with Society ethical requirements.