Physiology News Magazine
The International Mouse Phenotyping Consortium – the creation of a complete catalogue of mammalian gene function
News and Views
The International Mouse Phenotyping Consortium – the creation of a complete catalogue of mammalian gene function
News and Views
Steve Brown, Chair of the International Mouse Phenotyping Consortium & Director, MRC Harwell Institute, UK
https://doi.org/10.36866/pn.112.14

Despite the enormous advances in the characterisation of the human, mouse and other mammalian genomes, we remain remarkably ignorant of the function of the majority of genes. Much of the genome remains ‘dark’ and for many genes there is little or no functional knowledge. Moreover, while there have been advances in the identification of mutations in the human genome that lead to Mendelian Disorders and Rare Diseases, the identification of the genes and genetic variants that underlie common diseases has made slow progress. The functional characterisation of human genetic variation and its contribution to disease needs to be underpinned by a systematic effort to develop a complete catalogue of mammalian gene function. The International Mouse Phenotyping Consortium (IMPC) has risen to this challenge and in 2011 embarked on a global effort to identify the function of every gene in the mouse genome. The major mouse genetics centres from around the world have come together to develop the technologies and pipelines and build a genome-wide picture of gene function. It is expected that this catalogue will be transformative for biology and biomedical sciences providing a critical underpinning for our understanding of genes, genetic pathways and their impact on disease.
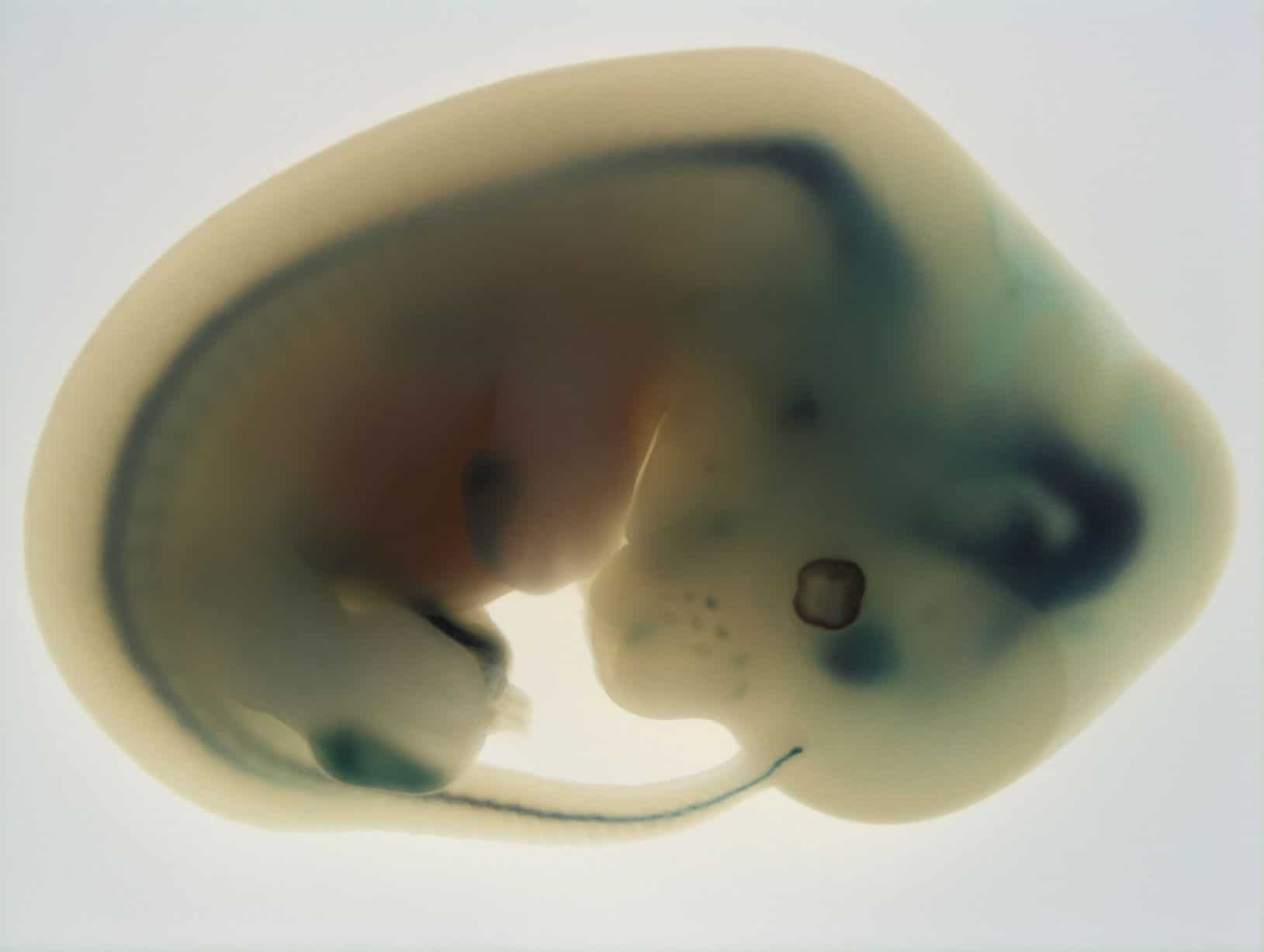
The IMPC is generating a null mutation for every gene in the mouse genome. Each mutant line undergoes a comprehensive phenotypic characterisation across many physiological and disease systems. Viable homozygous mutants enter an extensive and highly standardised adult phenotyping pipeline from 9 to 16 weeks of age. Homozygous lethal mutants enter an embryonic phenotyping pipeline which examines the timing of lethality and undertakes a detailed morphological analysis of embryological defects employing a number of sophisticated imaging platforms. From the generation of each mutant line to the acquisition of phenotyping data, considerable emphasis is placed on standardised procedures with stringent QC processes ensuring the development of a robust and reproducible dataset.
The IMPC comprises 19 research institutions and five national funders spread across 11 countries. 12 research institutions are principally responsible for mutant production and phenotyping and they deliver mutant and phenotype data to the IMPC Data Coordination Center (DCC) at the MRC Harwell Institute, UK. Following a stringent process of data wrangling and QC, data undergoes statistical analysis through the PhenStat tool to identify and assign phenotype calls to each mutant. All data is deposited at the IMPC core data archive at the EBI, Cambridge, UK, where it is made freely available through the IMPC portal. IMPC is an open access programme, and both the data and the mouse mutants are freely available to all academics. All the mouse lines are deposited in local archives and are available from IMPC and other national repositories around the world.
Since 2011, the IMPC has made substantive progress and has now analysed a third of the coding genome. It has generated over 7,500 mouse mutant lines, of which the majority have been completely phenotyped. This endeavour has to date generated a very large multidimensional dataset of genes and phenotypes comprising 62 million datapoints and 370,000 images. Already the IMPC programme is providing a series of extraordinary insights into the nature of the genetic landscape of the mammalian genome.
In 2017, the IMPC described a detailed analysis of the 410 homozygous lethal (essential) genes identified from the first 1,751 knockouts using a standardised phenotyping platform incorporating high-resolution 3D imaging. A large number of embryonic phenotypes were discovered for previously uncharacterised genes. An extraordinary and unexpected degree of variable expressivity was observed reflected in the large number of subviable mutations identified. Moreover, IMPC confirmed that human disease genes were significantly enriched for mouse essential genes. Building on this first major analysis of the genome landscape, IMPC undertook a comprehensive mouse-human cross-species analysis of data from Release 5.0 of the IMPC covering 3,328 genes. Ninety percent of gene-phenotype annotations had not been reported before and hitherto there had not been a mouse mutant available for 1,830 out of the 3,328 genes analysed. Moreover, for 1,092 genes the first functional knowledge was provided. Overall, the data demonstrate the huge fund of novel disease models that are being generated with wide implications for biomedical sciences and the major human and clinical genetics initiatives that are examining the genetic bases for disease, such as the 100,000 genomes project and the Precision Medicine Initiative. Moreover, the IMPC for the first time were able to take a comprehensive, genome-wide view of sexual dimorphism effects, particularly the interaction between genotype effects and sex. Extraordinarily, we found that for continuous parameters around one-sixth of phenotypes in mutants were affected by sex. Sexual dimorphism is pervasive across the mammalian genome, and it will be vital within the Precision Medicine Initiative and other programmes to take these findings into account. Finally, IMPC has focused on the identification of novel genes and pathways for diverse areas of physiology and disease, and two major reports have already appeared covering the auditory system and metabolism.
The IMPC has brought together the major mouse centres across the globe to tackle a major challenge for genetics and genomics and provide an increasingly comprehensive view of gene function and the role of genes in disease. The combined efforts and expertise of IMPC members have already generateda powerful dataset that is being used to generate transformative insights into the genome and disease landscape. The IMPC will continue its work over the coming years and complete the analysis of the coding genome. However, the consortium is already turning its attention to wider challenges as it continues to survey the need for functional genomic insights that will be critical in delivering a better understanding of the impact of human genetic variation on disease. A major priority will be to deliver an IMPC pipeline for the generation and characterisation of mice carrying human coding mutations. Such a pipeline will assist in the validation of putative human coding disease alleles as well as generating models for the pursuit of disease mechanisms and improved therapeutics. In addition, IMPC is considering plans to tackle a systematic effort to target and understand the function of the non-coding genome. One area of focus would be conserved non-coding enhancer elements (CNEs) which are highly enriched for human pathogenic variants in common diseases.
In conclusion, IMPC has created a highly productive international partnership to allow comprehensive functional analyses of the mouse genome with profound impacts on our understanding of the mammalian genetic landscape. IMPC will continue to address future challenges in the functional analysis of human genetic variation, aiming to bring further understanding and insight to this critical area of human disease studies, as well as uncovering opportunities for the development of therapeutic strategies.
Further Reading
Bowl MR et al. (2017). A large scale hearing loss screen reveals an extensive unexplored genetic landscape for auditory dysfunction. Nature Communications 8(1), 886.
Dickinson ME et al. (2016). High-throughput discovery of novel developmental phenotypes. Nature 537(7621), 508.
Karp NA et al. (2017). Prevalence of sexual dimorphism in mammalian phenotypic traits. Nature Communications 8, 15475.
Meehan TF et al. (2017). Disease model discovery from 3,328 gene knockouts by The International Mouse Phenotyping Consortium. Nature Genetics 49(8), 1231.
Oprea TI et al. (2018). Unexplored therapeutic opportunities in the human genome. Nature Reviews Drug Discovery 17(5), 317.
Rozman J et al. (2018). Identification of genetic elements in metabolism by high-throughput mouse phenotyping. Nature Communications 9(1), 288.
