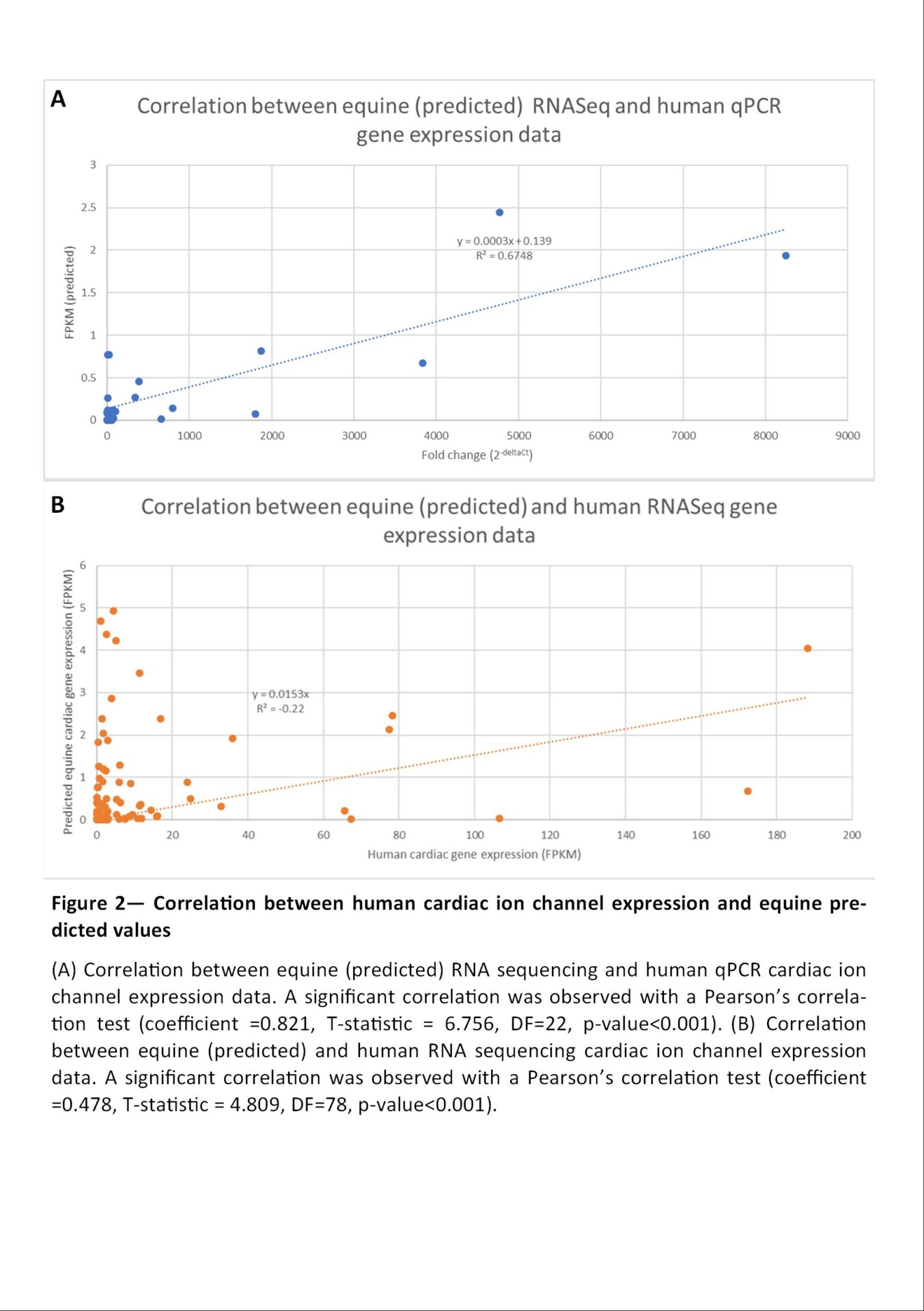
Neurons, muscle fibres and cardiomyocytes are the only three excitable cell types in any mammals. Their electrophysiologic properties are determined by specific proteins controlling the ion flux through the cellular membrane. These ion channels are essential for many physiologic and pathologic processes and constitute the targets for many drugs and are responsible for both their therapeutic and adverse effects. Knowing their expression across cardiac, cerebral and muscular tissue is the first step for understanding electrophysiological and pathological mechanisms and developing new treatments. Up to now, equine tissue-specific gene expression data is available for normal brain and muscle but not for the heart. Only one study sequenced a multi-tissue mix including cardiac tissue. Our work aims to predict the cardiac gene expression of ion channels based on open-source RNA sequencing data obtained from this multi-tissue mix. Material and methods Raw RNAseq data from normal horses from 9 datasets were retrieved from ArrayExpress and European Nucleotide Archive and reanalysed. These 9 datasets included RNA sequences from 13 tissues plus a mix of 43 tissues including cardiac muscle. Data was processed in R-Studio for quality control, alignment, read counts and data analysis using the Bioconductor packages RSubread, ShortRead. To predict the cardiac-specific gene expression, the normalized (FPKM) read counts Gik, for a gene i, in a sample k, was hypothesised to be the average of the expected expression in the tissue j αij weighted by the proportion of the tissue in the mix p (Figure 1, equation 1). The cardiac-specific expression was calculated by estimating the mean expression in each other tissues (Figure 1, equation 2). When the data was unavailable the median expression in all tissues was used. To test the validity of the model, predicted gene expression values were compared to the expression in human cardiac tissue determined by RNA sequencing. Results Ion channel expression exhibited tissue-specific expression patterns. Samples originating from the brain, muscle, uterus, kidney and testis showed the highest and most diverse expression of ion channels. The cardiac-specific expression could be predicted for 92 ion channels, calcium-handling proteins, pumps and exchangers. Comparison with human RNA sequencing (Ref1) and qPCR (Ref2) data showed a significant correlation with the experimental equine values (Pearson’s test of correlation, p-value<0.001) and with the predicted values after removal of the outliers (Figure 2, Pearson's test of correlation, p-value<0.001). Conclusion This work gives the first insight into the cardiac-specific ion channels expression in the horse and could serve as a base for targeting genes for future differential gene expression analysis.
Physiology 2021 (2021) Proc Physiol Soc 48, PC072
Poster Communications: Ion Channel Expression in the Equine Heart – In-silico prediction utilising RNA sequencing data from mixed tissue samples.
Antoine Premont1, Charlotte Edling1, Rebecca Lewis1, Kamalan Jeevaratnam1
1 Faculty of Health and Medical Sciences, University of Surrey, Guildford, United Kingdom
View other abstracts by:
Where applicable, experiments conform with Society ethical requirements.