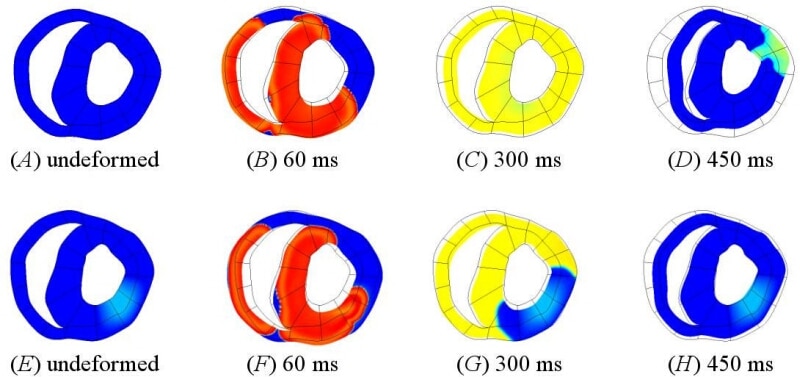
We have developed a dynamical model of pH regulation and acidosis in the myocyte. Acidosis of myocardium is correlated with reduced strength of muscle contraction (Orchard & Kentish, 1990), and disturbances to heart rhythm (Orchard & Cingolani, 1994). These effects are mediated through interactions between protons (intra- and extra-cellular) and various proteins in the myocyte, which couple the regulation of pH to ionic transport, and so also affect the cellular homeostasis of other ionic species. Based on data reported by Vaughan-Jones and colleagues (Leem et al. 1999), we developed multi-state enzyme-kinetic equations describing the four transmembrane proton or acid-equivalent ion transporters (Na+-H+ exchange, Na+-HCO3– cotransport, Cl–-HCO3– exchange and Cl–-OH– exchange) and the physicochemical buffering of pH in the myocyte. We have extended the existing modelling framework for myocyte electrophysiology, calcium handling and contraction to include the pH-dependence of key sarcolemmal ion channels and transporters involved in calcium handling and electro-mechanical coupling, in particular the inhibitory effects of protons on the L-type Ca2+ channel, the RyR Ca2+ release sites of the sarcoplasmic reticulum and the Ca2+ pump SERCA. The model is able to reproduce data on acid loading experiments (Leem et al. 1999), and predicts time courses for key ionic species during acidosis in the beating heart, in particular rising intracellular Na+ and Ca2+, and the inhibitory action of protons on the contractile proteins. We have embedded the cellular model of acidosis into a 2-dimensional slice (Crampin et al. 2004) of the Auckland heart model (Smith et al. 2004) to investigate altered contraction and pump function arising from region of acidosis (Fig. 1). The simulation shows an altered electrical activation sequence and reduced tissue deformation in the vicinity of the region of acidosis in the left ventricular wall. This study indicates that a computational modelling framework will provide a useful mechanism for investigating the effects of acidosis on heart function, and represents an important step towards a fully dynamic model of myocardial ischaemia.
University of Oxford (2004) J Physiol 561P, PC3
Communications: A DYNAMIC MODEL OF pH REGULATION IN THE MYOCYTE
Crampin,Edmund John; Smith,Nicolas Peter;
1. Bioengineering Institute, University of Auckland, Auckland, New Zealand.
View other abstracts by:
Figure 1. A tissue slice from the Auckland ventricular model (Smith et al. 2004) showing cell membrane potential and tissue deformation during normal electrical excitation (A)-(D) and with a region of acidosis in the left ventricular wall (E)-(H). The undeformed mesh is shown for reference.
Where applicable, experiments conform with Society ethical requirements.