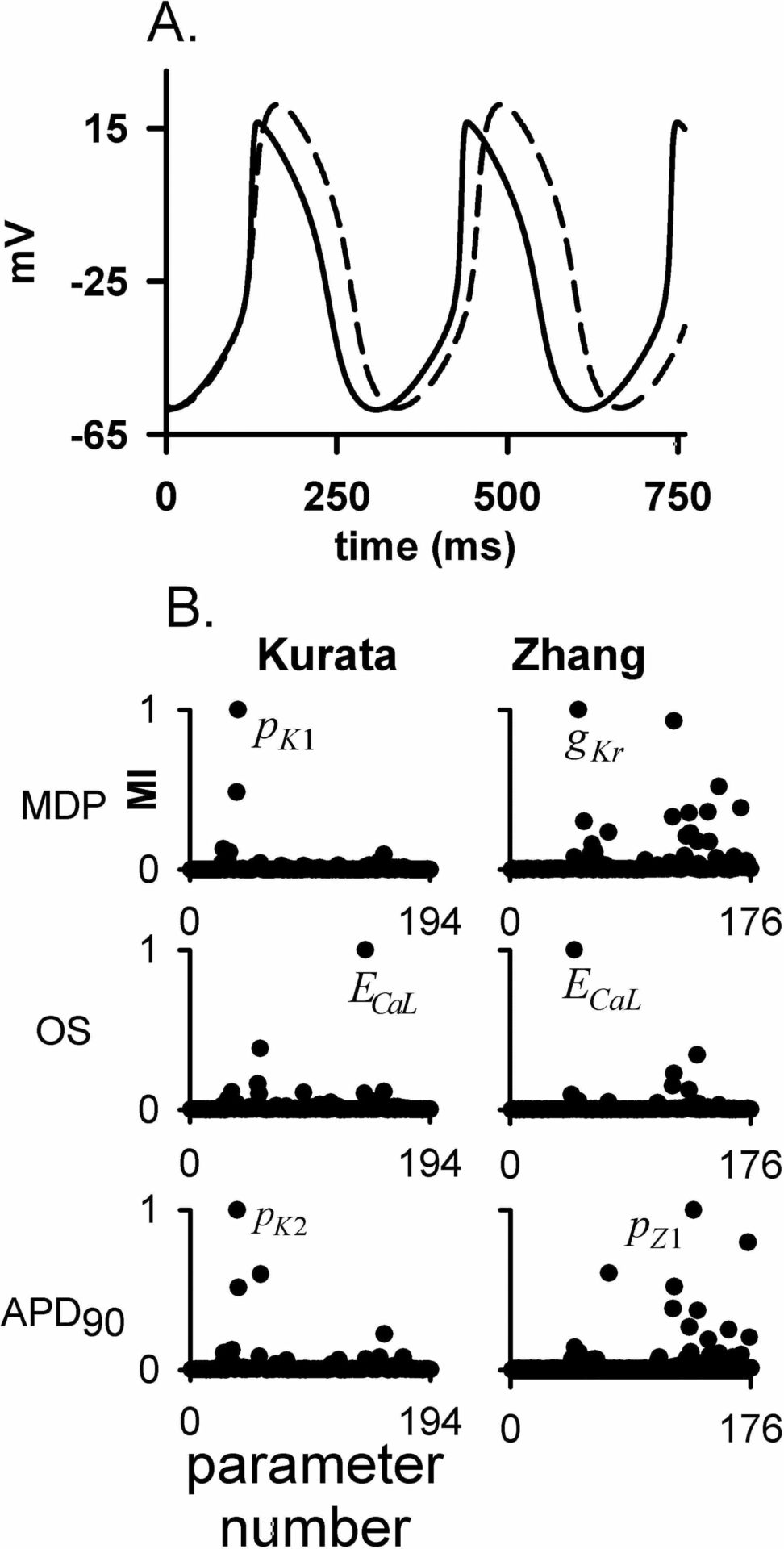
Aim: Biophysical cardiac cell models consist of coupled ordinary differential equations with many regulatory biophysical parameters (> 100). Model responses are differentially regulated by such model specific parameters. This study quantitatively compares the underlying prametric regulation of model responses among two rabbit pacemaker models using a systems biology approach with a novel global sensitivity mutual information index (MI). Methods: The rabbit pacemaker models developed by Kurata et al. [1] (Kurata) and Zhang et al. [2] (Zhang) were parameterically analysed in this study. Model responses in terms of action potential (AP) features were defined as maximum diastolic potential (MDP), over shoot potential (OS) and AP duration (APD90) [3]. Independent parameters associated with channel conductances, gating steady states, time constants and intracellular ionic homeostasis in the models were identified with 194 in the Kurata case, and 176 in Zhang case. The global information-theoretic sensitivity MI index has been developed in a previous study [4]. It constitutes deterministic quantification of the correlation between modelling parameters and model responses. Through multiple evaluations of the model (> 105) for randomly selected configurations of parameters, distirbutions of samples of model responses based on the perturbed parameter sets were obtained. With the large sampling, MI gave a quantitative statistical correlation between individual parameters and model responses. With such quantification, model parameters with respect to each model response were ranked from most relevant (MI = 1) to uninfluential (MI ~ 0). This enabled models comparision at parametric level. Results: Standard AP profiles from the models are shown in Fig. 1A. Due to the differing parametric regulation of the models, the two AP profiles are different. This is reflected in the differential parametric responses as seen in Fig 1B. Relative MI shows that MDP is mostly regulated by a component of the ICa,L channel activation time kinetics (pK1) in the Kurata case, whilst IK,r channel conductance (gKr) in the Zhang case. Reversal potential of ICa,L channel (ECa,L) regulates the OS in both models. APD90 is also regulated by differentially in the two models with a parameter of the ICa,L channel activation time kinetics (pK2) in the Kurata case and a parameter of the ICa,L channel inactivation time kinetics (pZ1) in the Zhang case. Conclusions: This MI analysis shows that although there is a common set of parameters regulating model responses, there are subtle differences. Often different models rely upon distinct mechanisms to reproduce various desired responses and MI provides a global stastical measure for identification of the same. MI can further assist in parameter estimation during model development.
University College Dublin (2009) Proc Physiol Soc 15, PC115
Poster Communications: A Systems Biology Computational Comparision of Two Caridac Pacemaker Cell Models
S. Kharche1, L. Ming2, H. Zhang1
1. School of Physics and Astronomy, University of Manchester, Manchester, United Kingdom. 2. School of Medicine, University of Manchester, Manchester, United Kingdom.
View other abstracts by:
(A) Standard AP profiles from Kurata (solid line) and Zhang (dashed line) models. (B) Left column show relative MI data for Kurata and right column for Zhang. Top panels show data for MDP, middle for OS and bottom for APD90. Parameters that maximally influence the respective model responses are shown. See text for details.
Where applicable, experiments conform with Society ethical requirements.