Intro. Napping is a common habit in many countries, but the impact of napping on general health is still unclear. Studies about the chronic effects of napping on obesity are contradictory, and the molecular link between napping and metabolic alterations has yet to be studied. Studies in animals and humans have shown that several genes expressed in adipose tissue (AT) follow a circadian rhythmic pattern, and disruption of circadian rhythms leads to obesity and metabolic disorders.
Aim. We aim to identify molecular mechanisms in AT that may connect napping and abdominal obesity. We propose that napping may induce, in AT, changes in the rhythmicity in those genes involved in energy metabolism, inflammation, and adipogenesis.
Method. We analyzed the circadian rhythmicity and global expression of genes expressed in 24-h cultured AT explants in 17 subjects with severe obesity, to assess differences between nappers (n=8) (long nappers, nap duration=1.18±0.7, mean±SD) and non-nappers (n=9). We extracted the RNA repeatedly across 24h from cultured AT explants of habitual nappers and non-nappers and performed RNA sequencing. Circadian rhythms were analyzed using 6 consecutive time points during 24 hours. Biostatistical and bioinformatic analyses were carried out in R. This work is in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki) and procured in line with World Health Organization Guiding Principles on Human Cell, Tissue and Organ Transplantation.
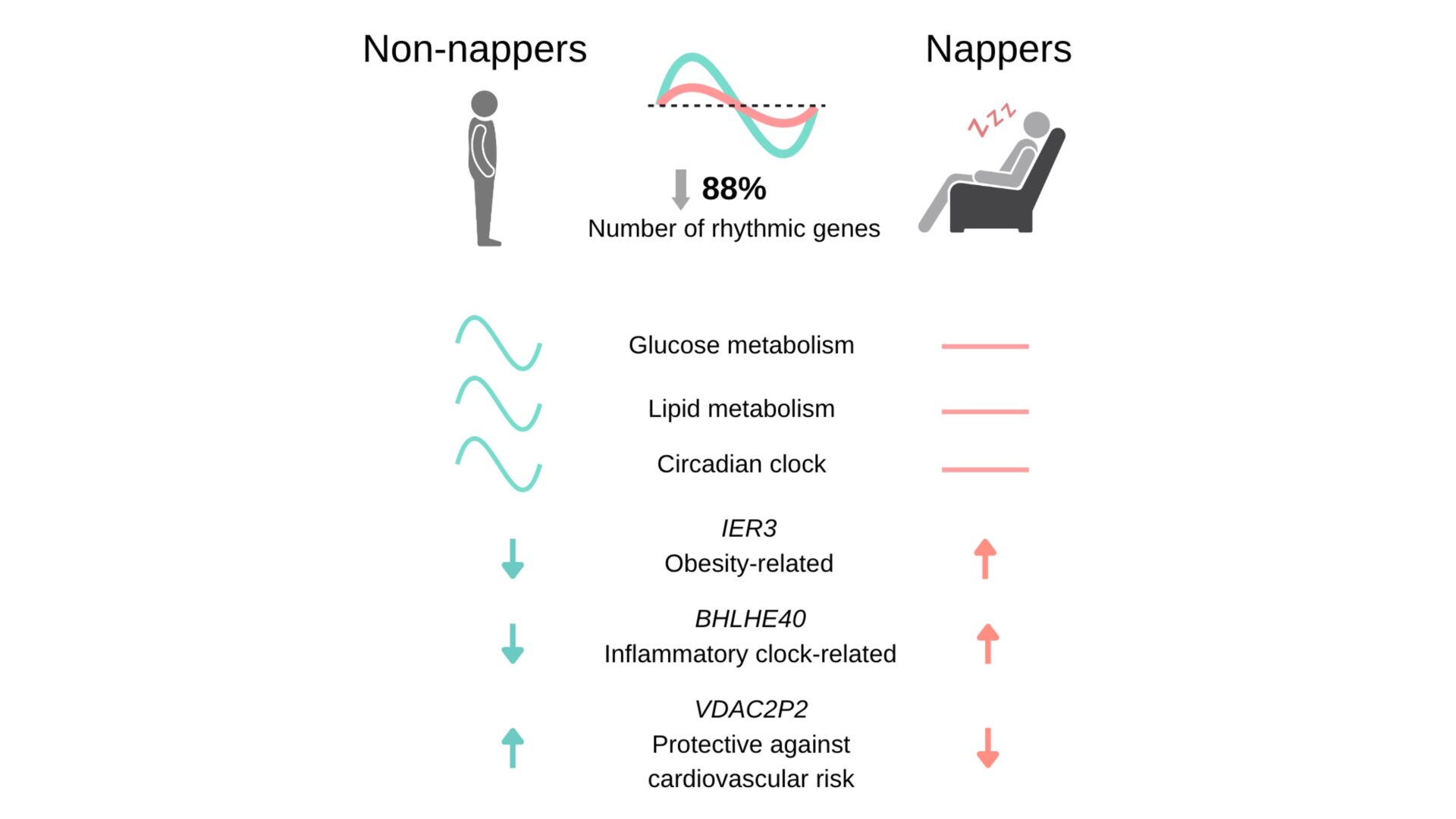
Results. RNAseq analysis showed that 28.31% of the total genes analyzed in the total population displayed significant rhythms both by cosinor and non-parametric analyses. We further divided the total population into nappers and non-nappers to answer our main aim. In nappers, there was a loss of rhythmicity in 88% of genes that showed circadian rhythmicity among non-nappers (among non-nappers, 2666 genes were rhythmic, while among nappers, only 321 genes showed circadian rhythmicity). We observed a reduction in rhythm amplitudes of 29% in nappers and identified cholecystokinin (CCK) as the gene with the strongest decrease in amplitude in nappers. We also found significant phase differences from a coherent unimodal acrophase in non-nappers towards a scattered and bimodal acrophase in nappers. CTNNAL1 and CHUK were two relevant genes with significant delay in acrophase. Further pathway enrichment analysis showed that those genes that lost rhythmicity with napping were mainly involved in pathways of glucose and lipid metabolism, as well as of the circadian clock. Additionally, we found differential global gene expression between nappers and non-nappers, with 34 genes down- and 32 genes up-regulated in nappers. The top-up-regulated gene (IER3), another gene highly up-regulated (BHLHE40, a clock-related gene), and top-down-regulated pseudogene (VDAC2P2) in nappers have been previously shown to be involved in inflammation.
Conclusion. These findings not only resolve conflicting evidence on napping's chronic effects on obesity but also offer crucial insights into molecular mechanisms, shaping our understanding of napping's role in metabolic disorders. Since napping during the day, especially long napping, may alter the AT circadian rhythm and this may influence obesity, we would recommend not taking long naps, based on our previous results.