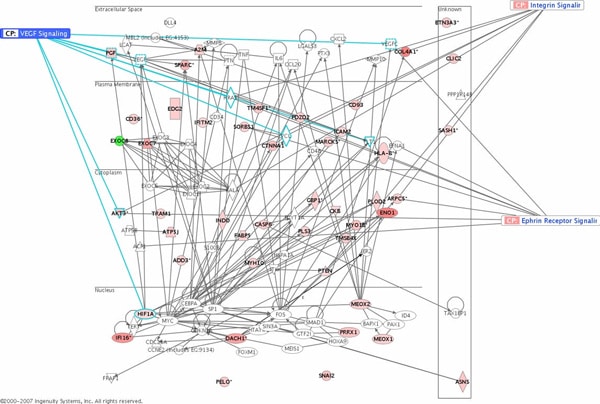
We have previously produced the first coding genome wide profile of the trancriptome responses to 6 weeks of endurance training in sedentary but otherwise healthy male subjects. This utilised a U95 Affymetrix gene chip platform and 8 subjects from 24 involved in the clinical study. Subsequent RT qPCR revealed that certain angiogenic and extracellular matrix (ECM) gene expression responses were modulated in proportion to physiological adaptation. This, for the first time, provide evidence that transcriptomic did not only catalogue changes with a stimulus but could reveal important gene networks likely to represent critical processes involved in physiological adpation. In the present study we have utilised the U133+2 Affymetrix genechip (n=48) in 24 subjects before and after 6 weeks of supervised endurance training (45min per day, at 70% VO2max). We discovered that 1100 transcripts are regulated 24hr post last training session, and these genes again reflected ECM processes, based on gene ontology classification. Further analysis revealed that 100 genes distinguish those that improved there aerobic function, from those that did not (20% of the cohort). Using the Ingenuity Database of hand-curated biological interactions we identified 5 prominent gene networks within this 100 gene list (Fig 1). Subsequent analysis of 30 of the 100 genes, in rodents bred for high and low training responsiveness, using real time qPCR identifies which of these marker genes are conserved from an evolutionary perspective. As these genes represent key factors relevant for adaptation to physical activity, it is also plausible that they represent “thirfty” genes, and further analysis, associating them with metabolic disease and diabetes is merited. In summary, we present the most detailed map of the molecular determinant of endurance training adaptability in mammals and identify the key signalling processes relevant for transducing physical activity into gains in aerobic capacity.
Life Sciences 2007 (2007) Proc Life Sciences, C36
Research Symposium: Defining the essential mammalian endurance training transcriptome
J. A. Timmons1, 2, N. Vollaard1, C. Sundberg2, C. Scheele2, 1, E. Jansson2, L. G. Koch3, C. Wahlestedt2
1. SLS, Heriot Watt University, Edinburgh, United Kingdom. 2. Centre for Genomics and Bioinformatics, Karolinska Institutet, Stockholm, SE171 77, Sweden. 3. Department of Physical Medicine and Rehabilitation, University of Michigan, Ann Arbor, , Michigan, MI, USA.
View other abstracts by:
Where applicable, experiments conform with Society ethical requirements.