Deep Brain Stimulation (DBS) is a common procedure in people with severe movement disorders – such as Parkinson’s Disease (PD), Essential Tremor (ET) and Dystonic Tremor (DT) – once symptoms are no longer manageable with medications. DBS consists in stimulating specific areas of the patient’s brain through an implanted electrode. However, stimulation approaches are still rudimentary. Reliable physiomarkers for discriminating neural response from the stimulation are needed to enable a closed-loop adaptive DBS system for personalised therapy.
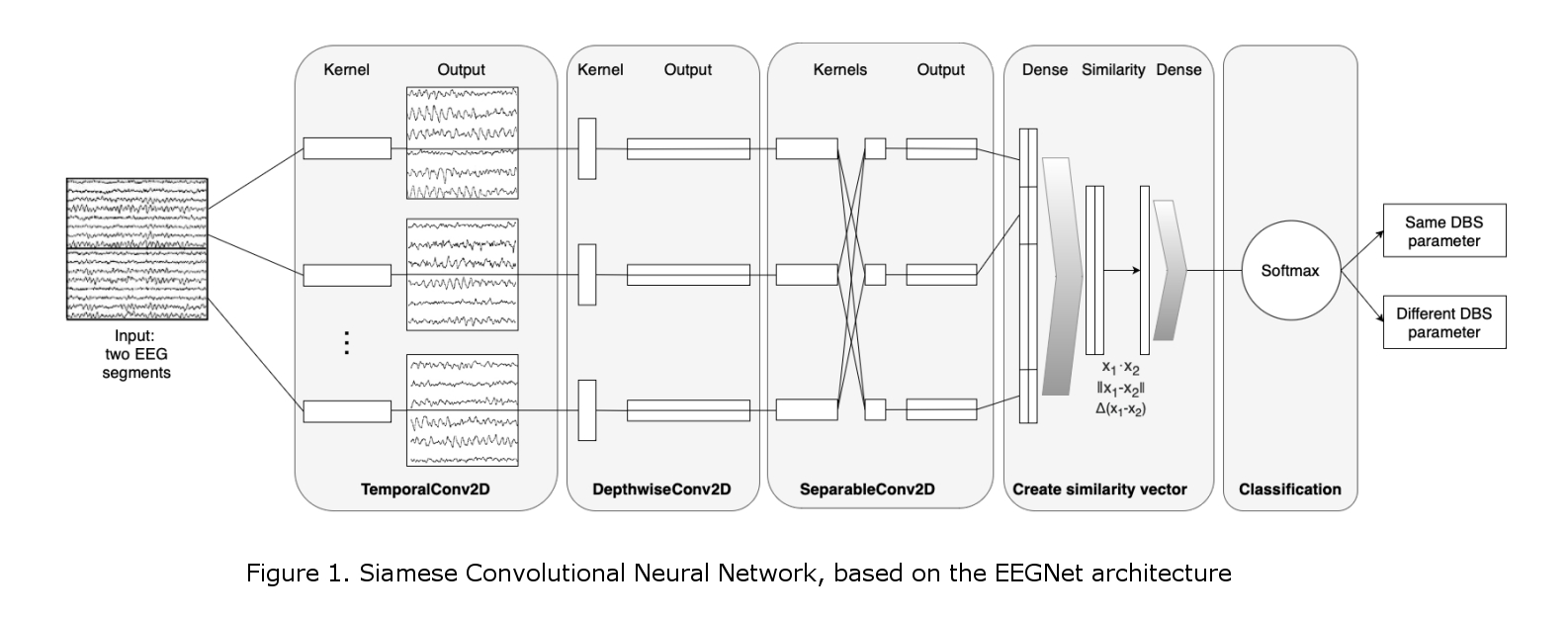
Here, a deep learning model for agnostic physiomarker search in EEG recordings is presented. Our Convolutional Neural Network (CNN) is based on the EEGNet structure [1], and our proof-of-concept work of classifying EEG activity during DBS [2]. Our Siamese CNN (Figure 1) was created for the EEG-based distinction of different DBS settings (same vs different). In addition, an explainability method based on ablation studies is proposed for identifying the spectral location of the features utilized by the model [2], which can be used as physiomarkers. All work presented here was conducted on a labelled EEG dataset of 1 DT and 1 PD patient recorded during their initial DBS monopolar review.
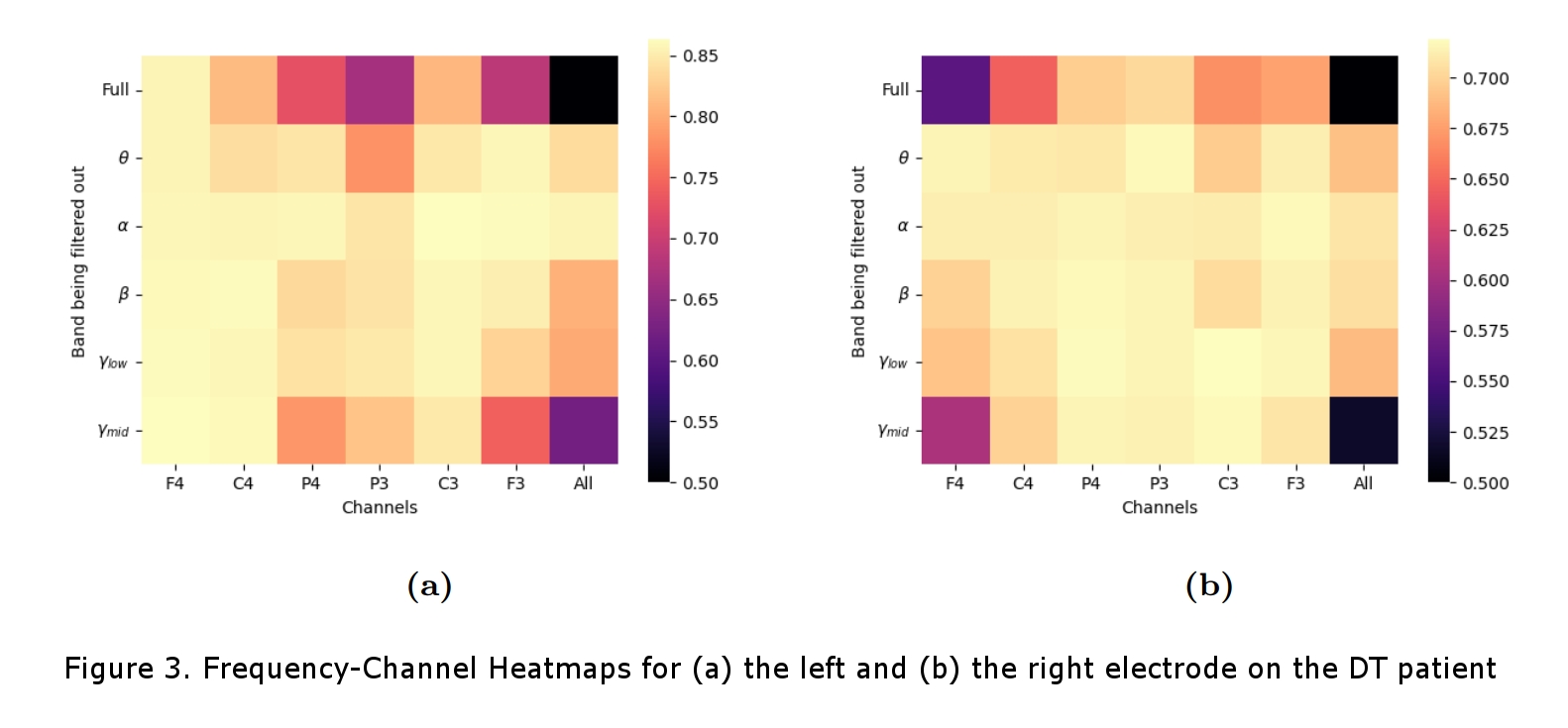
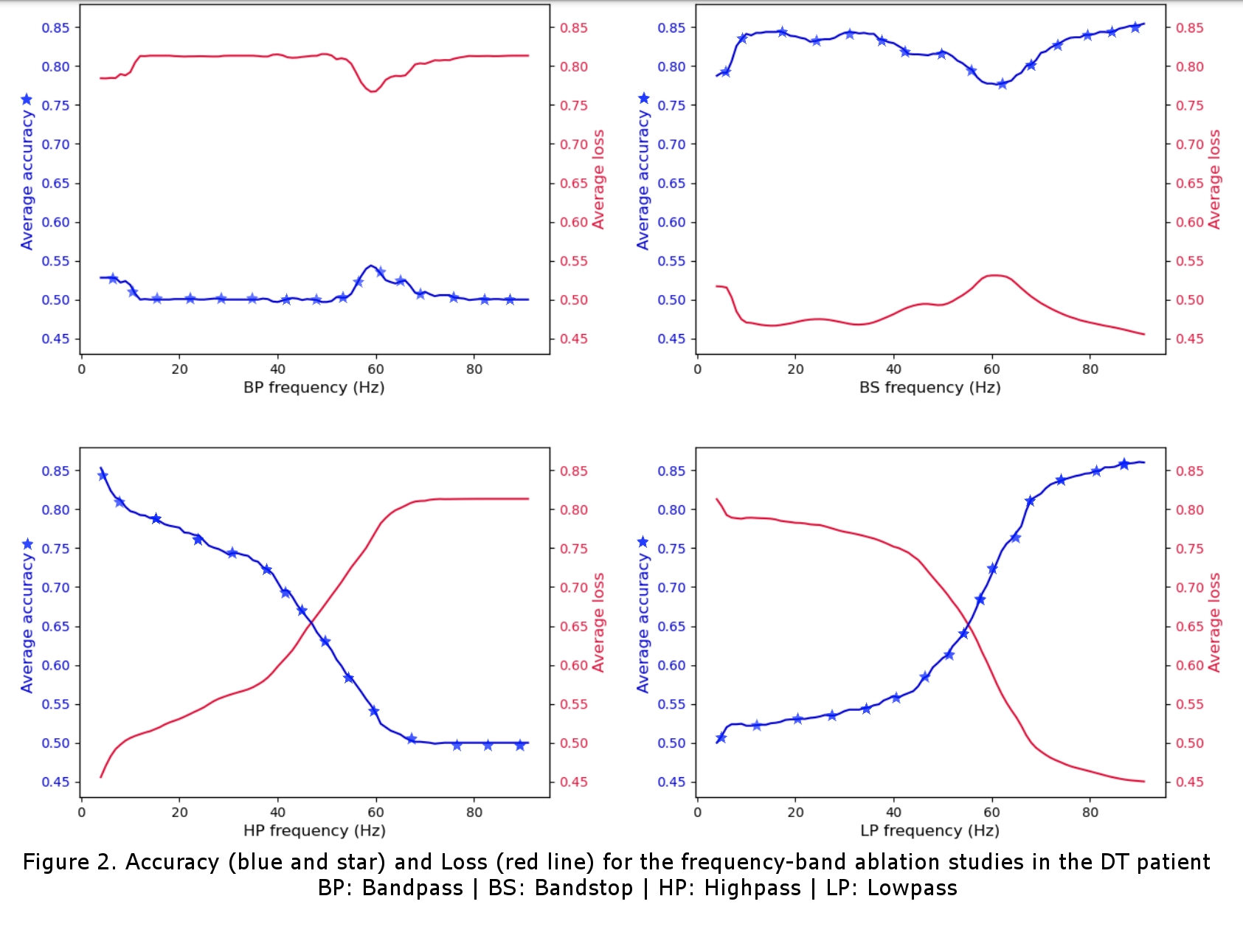
For the DT patient, cross-validation accuracies of 86% and 71% were achieved for the left and right contact respectively and, for the PD patient, accuracies of 75% and 63% were achieved. With the ablation studies (Figure 2), we found that, for both patients, features in the theta band (4-8Hz) and narrow-band gamma (NBG) band (60-90Hz) were the main contributors to the accurate classification of the data. The theta band was attributed to the patients’ tremor and the NBG band was identified as a potential physiomarker for DBS response in EEG data. The channel-wise ablation studies (Figure 3) revealed that, as expected, different channels were used for the classification task for the different DBS leads (left vs right), and overall, it showed that most channels were not used for the accurate classification of the stimulation.
The difference in accuracy between contact points was attributed to the monopolar review protocol which started with the left DBS and left it on while reviewing the right, presumably making it more difficult to classify changes in the stimulation from the right lead. It is also interesting to note that the DT patient in our study was not taking dopaminergic medicines, which’s intake has been recently seen as significant for detecting NBG physiomarkers [3]. Overall, our findings are in line with recent research in the field, which suggests the potential of the proposed approach in local field potential-based physiomarkers search [3, 4]. We have here provided a proof-of-concept of EEG-based DBS setting discrimination and have reaffirmed the recent shift in research from beta band (13-30Hz) physiomarkers to physiomarkers in the NBG range. Future work from our team will include a sample-size increase for statistical significance as well as making a patient-independent classifier with methods of transfer learning.