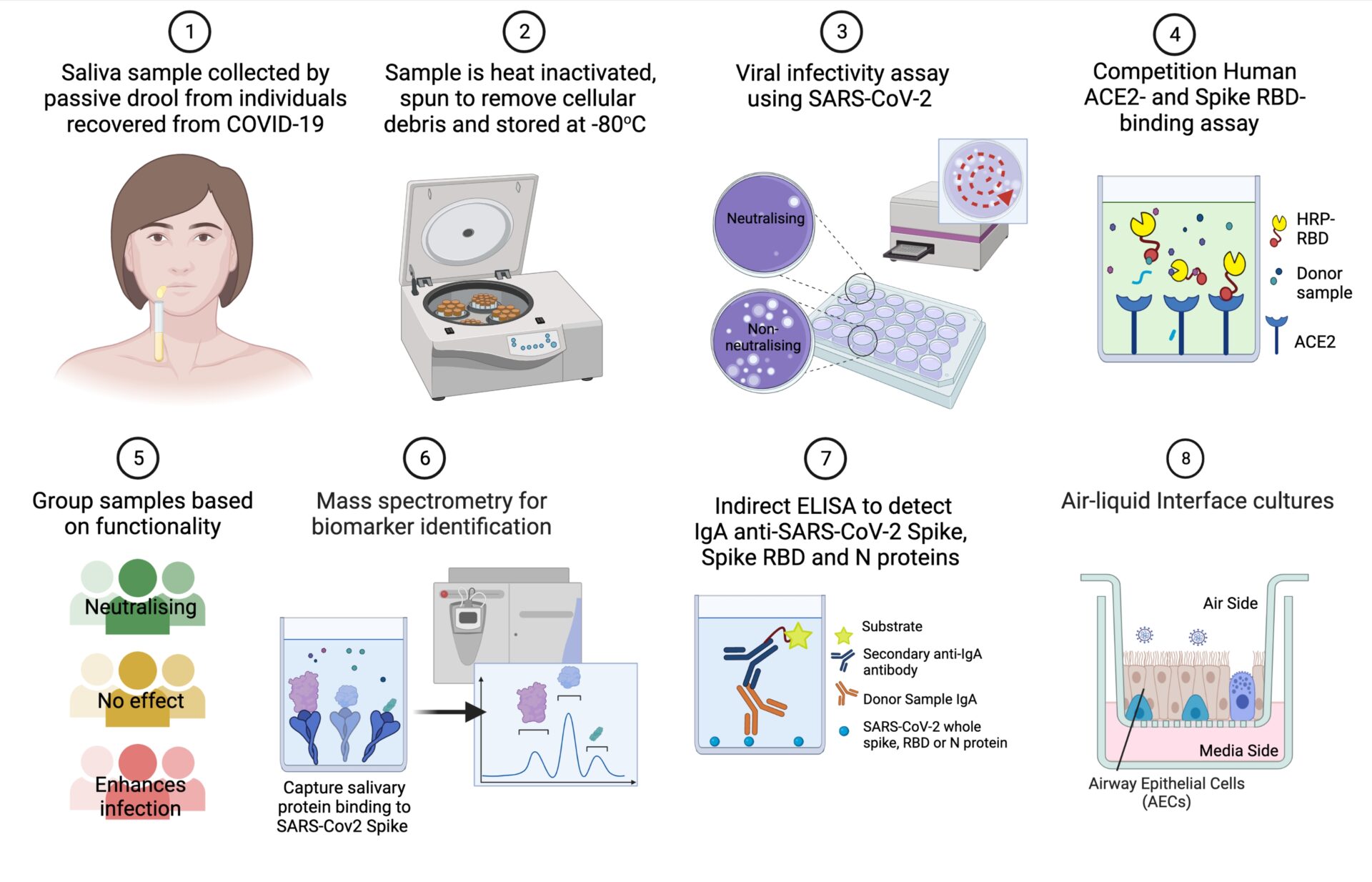
BACKGROUND: Our body's first defence against inhaled viruses like SARS-CoV-2 is a protective barrier that lines our respiratory tract called the mucosal-epithelial barrier. This barrier plays a vital role in neutralising the virus before it can infect respiratory epithelial cells. Secretions present in the nasal mucosa are also present in saliva, making it an ideal sample for non-invasive study of this barrier site. In this study, we aimed to identify key proteins found in saliva samples of healthcare workers who had recovered from COVID-19, which could protect against SARS-CoV-2.
METHODS: We collected 551 saliva samples from consenting staff at Great Ormond Street Children's Hospital who had previously tested positive for SARS-CoV-2 infection, prior to vaccination. Samples were grouped based on their functional ability to protect against infection using an in vitro RBD-ACE2 binding and infection assays with VeroE6 epithelial cells. A subset of samples was also tested for viral neutralisation using air-liquid interface (ALI) culture of primary human respiratory cells. The levels of proteins which specifically bind to SARS-CoV-2 spike antigen were measured by baited mass spectrometry and compared between the functional subgroups.
RESULTS: We found that 7.3% (n=29) of the screened saliva samples reduced SARS-CoV-2 infectivity >2-fold (Group A); 89.3% (n=353) had minimal effect on SARS-CoV-2 infectivity (Group B) and 3.3% (n=13) of samples resulted in >2-fold enhancement in SARS-CoV-2 infectivity (Group C). Proteomics analysis of samples from these subgroups (n=10 for each) identified elevated IgA in the most neutralising samples. This was supported by ELISA screening with IgA detected specific for the SARS-CoV-2 antigens Spike in 86% (422 of 488), Nucleocapsid in 85% (418 of 488), and RBD in 83% (377 of 450) of saliva samples. We determined that a salivary concentration of anti-RBD IgA above 500 pg/µg total protein significantly (p=0.035) reduced in vitro viral infectivity compared to saliva samples which tested negative for anti-RBD IgA.
Proteomics analysis also revealed elevated vimentin (VIM), antithrombin III, and S100A9 in the samples associated with enhanced SARS-CoV-2 infectivity (Group C). This was supported by further in vitro infectivity assays using recombinant VIM, SERPIN and S100A9 protein at concentrations corresponding to the detrimental saliva samples. This demonstrated that vimentin exposure resulted in the largest relative increase in SARS-CoV-2 infection of VeroE6 cells (22.2% increase versus mock). In addition, immunofluorescence staining of SARS-CoV-2 inoculated ALI cultures showed co-localisation of vimentin with SARS-CoV-2 antigen, indicating a role for vimentin in infection of human nasal epithelial cells.
CONCLUSION: Our research suggests that salivary IgA is a useful indicator of recent SARS-CoV-2 infection and higher levels of anti-RBD IgA can help reduce viral infection of epithelial cells. However, other secreted proteins were found to be associated with enhancing in vitro SARS-CoV-2 infectivity. Screening saliva for mucosal biomarkers such as vimentin may be an effective strategy to help identify individuals who are most vulnerable to repeat infection by SARS-CoV-2.