Background: Caffeine, found in coffee, tea, energy drinks, and some foods and supplements, is the most widely consumed psychostimulant in the world. Caffeine pharmacokinetics and pharmacodynamics vary between individuals (Nehlig, 2018), with single-nucleotide polymorphisms (SNPs) in the genes that encode for the adenosine receptor (ADORA2A) and the P450 enzyme (responsible for 95% of the body’s caffeine metabolism; CYP1A2) possibly responsible for some inter-individual variability. The purpose of this study was to determine the influence of two commonly occurring ADORA2A and CYP1A2 SNPs on caffeine metabolism.
Methods: Sixteen healthy adults (age 24 ± 5 years; BMI 22.9 ± 2.4 kg∙m-2; n = 3 women) volunteered to participate and consumed a capsule containing 3 mg∙kg-1 body mass of caffeine, after an overnight fast and having abstained from caffeine consumption for 48 h. Venous blood samples were collected pre, and 30 and 120-min post caffeine ingestion. Serum caffeine and paraxanthine were measured using high-performance liquid chromatography. Genomic DNA was extracted from whole blood and SNPs in ADORA2A (rs5751876) and CYP1A2 (rs762551) genes were determined by rhAmp assays (Integrated DNA Technologies, USA). Participants were categorised by ADORA2A gene as TT homozygous (‘high’ sensitivity) or C allele carriers (CT heterozygous or CC homozygous: ‘low’ sensitivity); and by CYP1A2 gene as AA homozygous (‘fast’ metabolisers) or C allele carriers (AC or CC: ‘slow’ metabolisers). Mixed model ANOVAs were used to examine serum caffeine, paraxanthine, and paraxanthine:caffeine ratio.
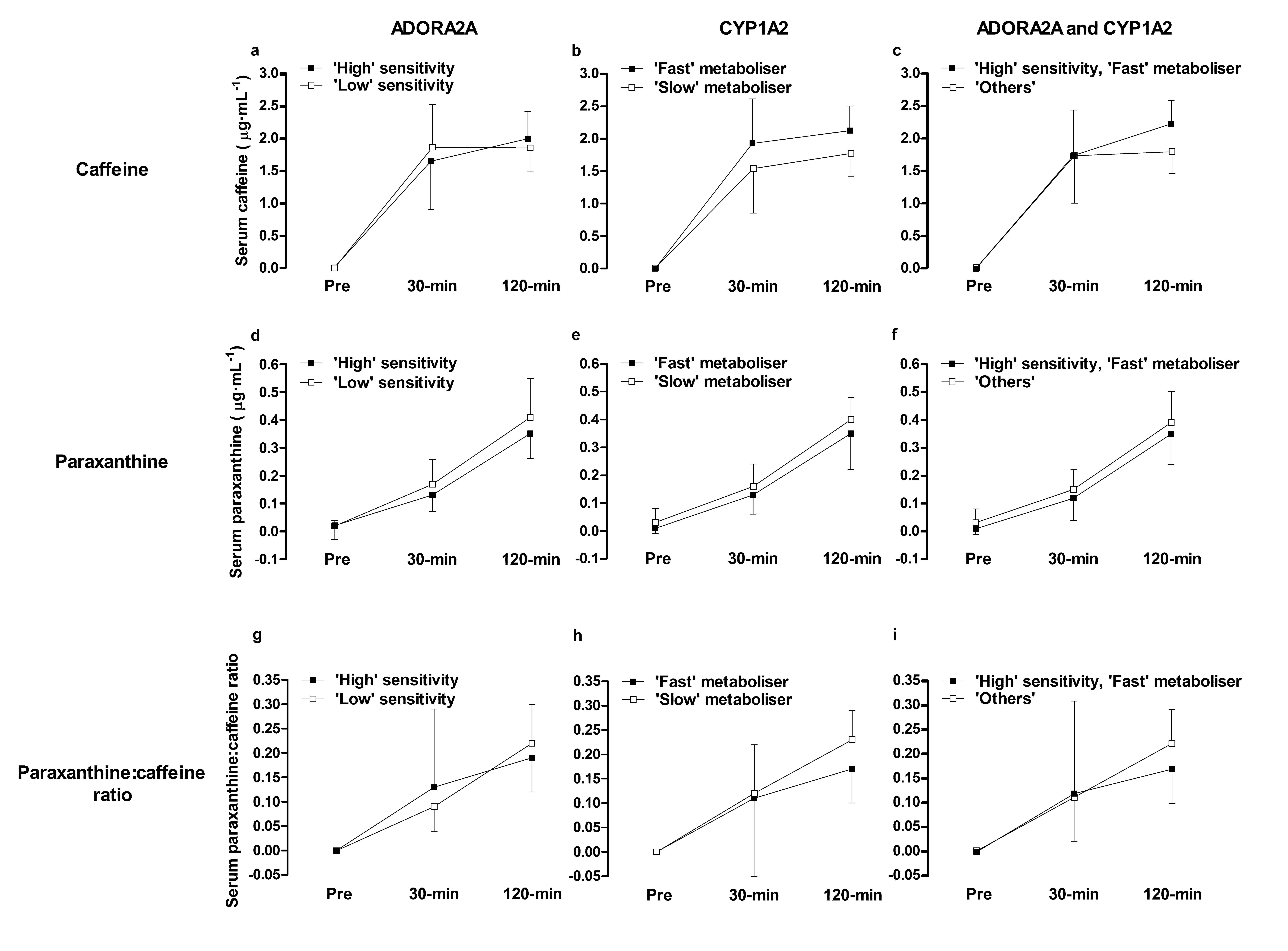
Results: n = 10 participants had ‘high’ and n = 6 ‘low’ sensitivity ADORA2A genotype; n = 8 had ‘fast’ and n = 8 ‘slow’ metabolism CYP1A2 genotype; and n = 6 had both ‘high’ sensitivity and ‘fast’ metabolism genotypes (i.e., ADORA2A, TT; CYP1A2, AA). There were no genotype x time interactions for serum caffeine (P ≥ 0.311), paraxanthine (P ≥ 0.486), or paraxanthine:caffeine ratio (P ≥ 0.433; Figure 1). Main effects of time were found for serum caffeine, paraxanthine, and paraxanthine:caffeine ratio (P < 0.001). Bonferroni corrected post-hoc t-tests revealed serum caffeine increased from pre-ingestion by 1.73 ± 0.69 and 1.94 ± 0.40 μg∙mL-1, at 30 and 120-min post-ingestion (P < 0.001). Serum paraxanthine increased from pre-ingestion by 0.12 ± 0.08 and 0.35 ± 0.10 μg∙mL-1, at 30 and 120-min post-ingestion (P < 0.001). Serum paraxanthine:caffeine ratio increased from pre-ingestion by 0.11 ± 0.13 and 0.20 ± 0.07, at 30 and 120-min post-ingestion (P = 0.01 and P < 0.001). There were no main effects of genotype for serum caffeine (P ≥ 0.07), paraxanthine (P ≥ 0.250), or paraxanthine:caffeine ratio (P ≥ 0.379).
Conclusion: Caffeine metabolism 30 and 120-min post caffeine ingestion was not different between healthy adults categorised by ADORA2A or CYP1A2 SNP genotypes. Responses during this period after ingestion may be more influenced by absorption. Additional study is warranted with longer monitoring periods (up to 6 h post-ingestion) to further examine metabolism responses. An influence of ADORA2A and CYP1A2 SNPs on some ergogenic effects of caffeine have been demonstrated previously (Grgic et al., 2021), but their influence on other physiological effects of caffeine require further examination.