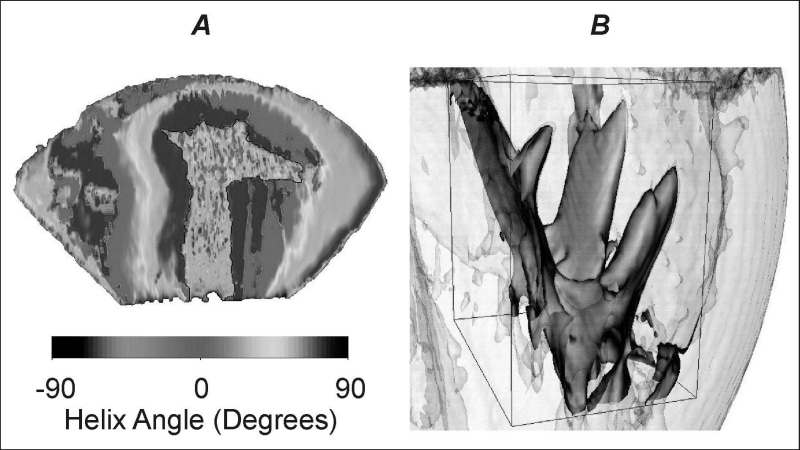
We reconstruct and visualise cardiac geometry and fibre architecture of post mortem canine hearts from diffusion tensor magnetic resonance imaging (DT-MRI) data sets. DT-MRI calculates 3 eigenvalues and 3 eigenvectors for the diffusion tensor for protons at each pixel throughout 3-dimensional tissue. The eigenvalues can be used to calculate a normalised measure of anisotropy: the primary eigenvector provides a measure of fibre orientation, while the tertiary eigenvector provides an index of sheet structure and orthotropy (Hsu et al. 1998). We visualise these DT-MRI tensors and extract the fibre orientation throughout the ventricles from the primary eigenvalues, or a combination of the linear and planar components of the tensor, with the 3D geometry of the ventricles reconstructed by stacking DT-MRI data slices. Fibre orientation in the left ventricular free wall, given by the helix angle of the primary eigenvector, changes smoothly from approximately −60 deg at the epicardial surface to 70 deg at the endocardial surface (Figure 1A). The image matrix for the canine ventricular data is 256 × 128 × 128, representing a physical field of view of 100 × 100 × 76.8 mm; each pixel in the DT-MRI data slices is 780 μm square, sufficient to visualise the boundaries of the papillary muscles and their insertions (Figure 1B), which are blurred by the temporal averaging of in vivo MRI of the beating heart. All stages of the model construction have been algorithmically automated: hence, DT-MRI of isolated hearts provides a high throughput method of reconstruction of detailed ventricular geometry and architecture that can be used for the computation of electrophysiology (Aslanidi et al. 2005).
University of Bristol (2005) J Physiol 567P, PC11
Poster Communications: Reconstruction and visualisation of cardiac ventricular geometry and architecture from diffusion tensor magnetic resonance imaging
Benson, Alan P; Li, Pan; Hsu, Edward W; Holden, Arun V;
1. Computational Biology Laboratory, School of Biomedical Sciences, University of Leeds, Leeds, United Kingdom. 2. Department of Biomedical Engineering, Duke University, Durham, NC, USA.
View other abstracts by:
Figure 1. A 3-dimensional reconstruction of a section of canine left ventricle showing a smooth transmural change in the helix angle of the primary eigenvector an indicator of fibre orientation. B the resolution of the DT-MRI data sets is sufficient to allow visualisation of papillary muscle.
Where applicable, experiments conform with Society ethical requirements.