Physiology News Magazine
The (concise) guides to pharmacology and what they provide for physiologists
Features
The (concise) guides to pharmacology and what they provide for physiologists
Features

Dr Steve Alexander
University of Nottingham Medical School, UK
A colleague suggested that the major difference between physiologists and pharmacologists is that the former mostly use drugs with names, while the latter mostly use drugs with numbers. The point (I think) they were making was that there is a greater chance that physiologists work with more defined systems and signalling pathways than pharmacologists. I don’t believe that to be true now, if ever it was. I would contend that a difference between the disciplines of physiology and pharmacology, while there are enormous overlaps, is that there is a wider use of concentration– and dose–response curves in pharmacology journals compared to physiology journals. (As is true of any generalisation, of course, there are always multiple examples of opposing evidence.)
The Guide to Pharmacology database and NC-IUPHAR
What the GuidetoPharmacology.org online database and the associated Concise Guides to Pharmacology aim to do is to make life easier for ANY scientist making use of drugs. The Nomenclature and Standards Committee of the International Union of Basic and Clinical Pharmacology (NCIUPHAR), which was established in 1987 and which I’ve chaired since 2015, has more than 100 subcommittees responsible for providing consensus information on families of pharmacological targets: receptors, transporters, enzymes and ion channels. Since the inception of NC-IUPHAR, close to 1,000 altruistic scientists worldwide have contributed time and information to the database, via a team of curators based at the University of Edinburgh, UK. The information the subcommittees collate includes rational, systematic nomenclature on their family of drug targets, based on a system initially defined for receptors (Vanhoutte et al.,1996). This ensures that biological scientists are all using the same language and avoids confusion. They also provide data on the characteristics of the distinct molecular targets, which might be biochemical, biophysical or pharmacological. For ion channels, for example, associated or ancillary proteins could be identified, as well as ion selectivities and voltage dependence for activation/inactivation.
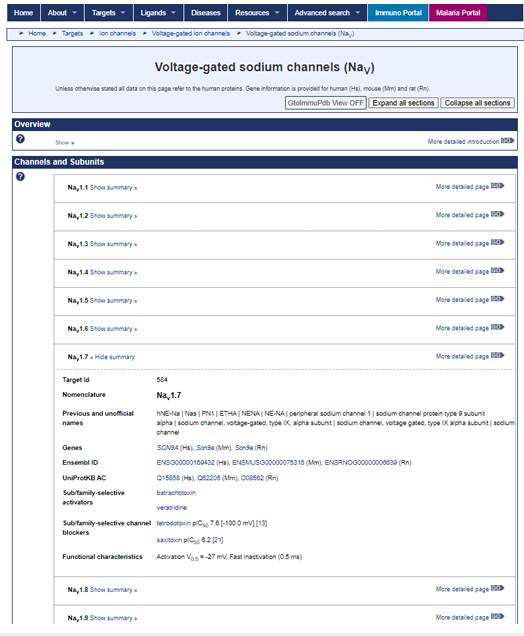
A key example of the contribution of an NC-IUPHAR subcommittee is the orderly naming of the voltage-gated ion channels. Based on a previous consideration of potassium channel nomenclature (Chandy, 1991), Bill Catterall and colleagues identified commonalities of structure of mammalian and non-mammalian channels, sufficient to describe a “chanome” superfamily of voltage-gated ion channels encoded by at least 143 genes in humans. A widely observed feature of the families of voltagegated ion channels was a selectivity for a single ionic species, with some variation in the mechanisms of endogenous regulation. In a 2003 series of publications in Pharmacological Reviews, introduced by Catterall et al., the subcommittee proposed the adoption of a systematic nomenclature naming individual voltage-gated ion channels using the chemical symbol of the principal permeating ion (Na, K or Ca) with the principal physiological regulator indicated as a subscript. Thus voltagegated sodium channels have the general description of NaV , while calcium-activated potassium channels are described as KCa. A numerical suffix identifies the gene family, while a number following a decimal point defines an individual channel isoform (e.g. NaV 1.1), where the numbering is organised chronologically. Many of the targets listed have both summary (Fig.1) and detailed views, where the former enables a more comparative approach and the latter a deep dive into an individual target’s properties. On the detailed view of NaV 1.7 on our database, we identify molecular and biophysical characteristics of the ion channels, linking to a multitude of additional online databases. We focus on human proteins primarily, with descriptions of mouse and rat orthologues, where these are available. Pharmacological tools listed distinguish activators, inhibitors, gating inhibitors and channel blockers, all with quantitative data, where appropriate, with a major emphasis on the evidence base of the literature. This highlights a second aim of the NC-IUPHAR subcommittees, which is the recognition of preferred tools for target identification. Clearly, we are not yet at the stage when every drug target can be defined by exceptionally selective tool compounds. NC-IUPHAR considered the situation almost 20 years ago – “When using drugs to define receptors or signalling pathways, it would be desirable to use a drug that acts only on the receptor or biological site of interest at all concentrations and doesn’t interact with others at any achievable concentration. Unfortunately, there are very few or no drugs with this ideal property. Fortunately, there are numerous drugs with a detectable potency difference (in exceptional cases >103-fold but usually much less) between their primary target and other related receptors.” (Neubig et al., 2003). The aim of the Guide is to provide guidance not only for which compound to use to define (as best as is possible) a particular ion channel, receptor, transporter or enzyme in a complex mix, but which concentrations of that agent will be most useful.
Concise Guides to Pharmacology
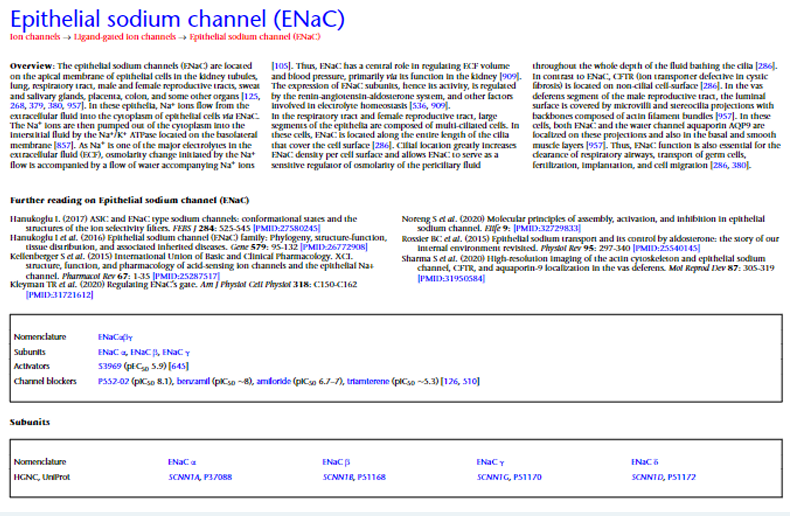
GuidetoPharmacology.org provides lists of agonists and antagonists for the receptors, with activators and inhibitors for the enzymes, ion channels and transporters. As of the start of 2022, these aggregate to 3,000 drug targets, with the inclusion of atypical “other” proteins. Lists of the 11,000 ligands are accessible as approved drugs, natural products, metabolites and synthetic organic compounds, as well as endogenous and other peptides, and antibodies. There are nearly 19,000 curated binding constants, with over 31,000 gleaned from large-scale screens and over 41,000 references. Such an abundance of information can be overwhelming, so the Concise Guide to Pharmacology takes a more pragmatic approach. While the online database presents (often very long) lists of information, the Concise Guide has a tabular, comparative approach (Fig.2) with a limited choice of fields and a restricted number of ligands described. It’s this approach that makes the Concise Guide concise, not the size of the Guide (the most recent version has 515 pages). The Concise Guide to PHARMACOLOGY 2021/2022 presents ~1,900 drug targets arranged in seven articles focused on ion channels, G proteincoupled receptors, nuclear hormone receptors, catalytic receptors, enzymes, transporters and other proteins (Alexander et al., 2021). Families of these drug targets are arranged in side-by-side comparisons identifying the preferred nomenclature, links to protein/gene databases, ligands (agonists, antagonists, inhibitors, antibodies) and their characteristics, including affinity/potency estimates, together with general comments. In a divergence from the online database, the priority in the Concise Guide is to feature ligands that are not just selective for the individual target (where appropriate), but are available either commercially or as a gift for researchers to employ.
One of the overarching aims of the Guide and the Concise Guide is to provide an evidence base for researchers to employ. This means that scientists new to a research area can confirm the appropriate compounds and concentrations to employ in complex mixtures to allow identification of individual drug targets. In addition, recommended further reading identifies suitable reviews for researchers to gain a broader contextual understanding. As such, the Concise Guides and the online database at GuidetoPharmacology.org provide a valuable resource for the community of physiologists, as well as pharmacologists. For electrophysiologists, in particular, the capacity to compare experimental observations of ion fluxes and drug responses to the information provided on GuidetoPharmacology.org should allow rapid identification of a particular channel (or suggest a novel entity). The most recent “point-in-time”, citeable update, The Concise Guide to PHARMACOLOGY 2021/2022 was published in the British Journal of Pharmacology in Autumn 2021 and is available online as seven freely downloadable pdfs.
Maintaining the currency and the quality
One of the features of working in the biological sciences is the pace of change. While experimental protocols and reagents from over a century ago may still be useful in chemistry, for example, the same is rarely true in the biological sciences. A feature of the Guide, and consequently the Concise Guides, is the contemporary nature of the material we present. We have a regular cycle of updates to coincide with the publication of the Concise Guides, where the subcommittees of NC-IUPHAR and other expert consultants provide their reflections on any changes to be incorporated into the database. With this in mind, we are contactable through the database website or via enquiries@guidetopharmacology.org should you identify an area that you think might be suitable for updating.
References
Alexander SP et al., (2021). The Concise Guide to PHARMACOLOGY 2021/2022: Introduction and Other Protein Targets. British Journal of Pharmacology 178 (Suppl 1), S1–S26. https://doi.org/10.1111/ bph.15537.
Catterall WA et al. (2003). International Union of Pharmacology: Approaches to the nomenclature of voltage-gated ion channels. Pharmacological Reviews 55, 573–574. https://doi.org/10.1124/pr.55.4.5
Chandy KG (1991). Simplified gene nomenclature. Nature 352, 26.
Neubig RR et al., (2003). International Union of Pharmacology Committee on Receptor Nomenclature and Drug Classification. XXXVIII. Update on terms and symbols in quantitative pharmacology. Pharmacological Reviews 55, 597–606. https://doi.org/10.1124/ pr.55.4.4.
Vanhoutte et al. (1996). X. International Union of Pharmacology recommendations for nomenclature of new receptor subtypes. Pharmacological Reviews 48, 1–2. https://doi.org/10.1124/pr.114.008862.
