Physiology News Magazine
Understanding skeletal muscle hypertrophy: integration of cell signalling
All physiologists know that muscle activity is necessary to maintain muscle mass. But what are the underlying molecular mechanisms at work in the muscle? Here Douglas Bolster and colleagues offer some insights
Features
Understanding skeletal muscle hypertrophy: integration of cell signalling
All physiologists know that muscle activity is necessary to maintain muscle mass. But what are the underlying molecular mechanisms at work in the muscle? Here Douglas Bolster and colleagues offer some insights
Features
Douglas R Bolster (1), Neil Kubica (1), Stephen J Crozier (1), David L Williamson (1), Peter A Farrell (2), Scot R Kimball (1) and Leonard S Jefferson (1)
1: The Pennsylvania State University College of Medicine, Department of Cellular and Molecular Physiology, Hershey, PA, USA
2: East Carolina University, Department of Exercise and Sport Science, Greenville, NC, USA
https://doi.org/10.36866/pn.55.18


The vital importance of skeletal muscle to general health and daily activities is likely taken for granted by most individuals. Skeletal muscle is essential for basic posture, movement, and a variety of metabolic functions. Specifically, skeletal muscle accounts for approximately 40 – 50% of the total body mass, serves as the predominant site for glucose metabolism and greatly contributes to the basal metabolic rate. Maintaining or even enhancing skeletal muscle mass becomes critical in the context of aging (sarcopenia) and various disease states associated with muscle loss (e.g. sepsis, cancer, diabetes, HIV). Thus, considerable efforts have been made recently to elucidate the cellular and molecular mechanisms by which skeletal muscle loss (atrophy) and gain (hypertrophy) occur. Importantly, the processes that invoke muscle atrophy are relatively unique and do not appear to be the simple reverse of hypertrophy.
Increases in skeletal muscle mass are dictated through the process of protein turnover, which is the balance between protein synthesis and protein breakdown. Higher rates of protein synthesis relative to protein degradation must be maintained in order to achieve hypertrophy, whereas elevated protein breakdown will induce a loss of protein. Overall, these processes continuously operate and are susceptible to external modulation by factors such as nutrient availability, hormones, and exercise.
Molecular regulation of growth
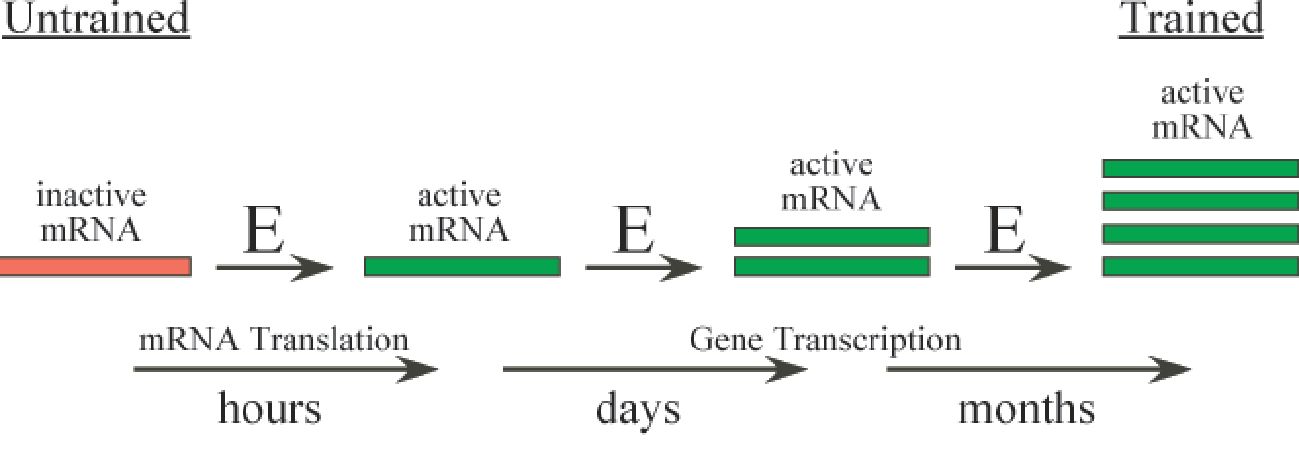
The molecular controls that govern changes in protein synthesis and eventual gain in muscle mass incorporate both transcriptional and translational inputs. Although exceptions occur, consequences associated with altered gene transcription generally occur over a period of days to weeks, whereas effects attributed to mRNA translation (i.e. the process of synthesizing a protein based on the information encoded by the mRNA) can be manifested within minutes to hours. Transcription and translation each contain three distinct steps (initiation, elongation, termination) with the predominant regulation at the phase of initiation. However, translation is unique because mRNA is recruited rather than produced and this process is responsive to acute metabolic/nutritional alterations. The focus of the present article will be to highlight the impact of mRNA translation initiation on acute changes in protein synthesis, the upregulation of select mRNAs related to growth and how these events culminate to alter gene expression in the context of resistance exercise.
Translation initiation and resistance exercise
Translation initiation essentially encompasses two central components mediated by eukaryotic initiation factors (eIFs) that control rate-limiting events. These two components in simple terms allow the ribosome to bind to the mRNA (eIF4F complex) and to bring the ribosome to the site on the mRNA where translation begins (eIF2/eIF2B)(Fig. 1). An essential mechanism for regulating growth within translation initiation involves the mammalian ‘target of rapamycin’ (mTOR) protein. Two common downstream targets of mTOR are the 70-kDa ribosomal protein S6 kinase (S6K1) and the eIF4E-binding protein-1 (4E-BP1).
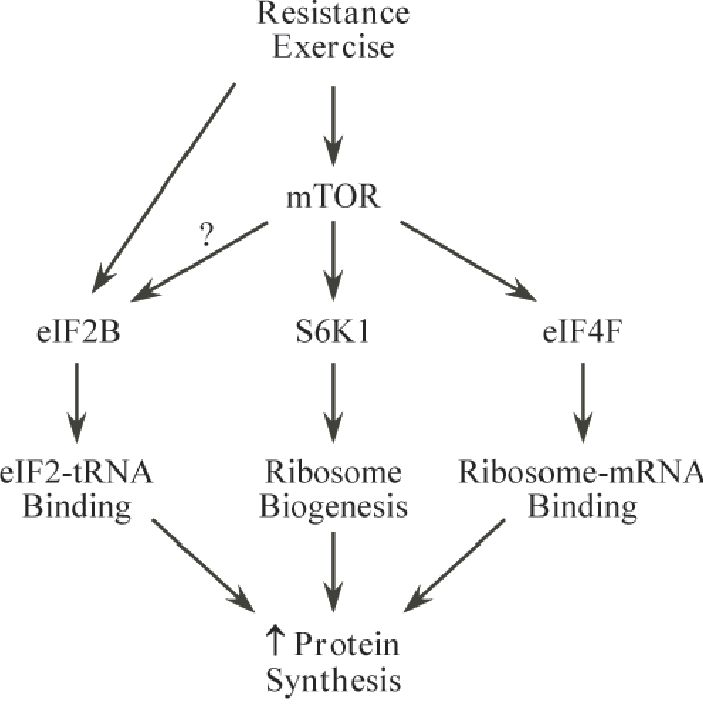
A common misconception regarding changes in translation initiation is that activation of any protein in this pathway corresponds with increases in protein synthesis. For instance, following resistance exercise, elevations in protein synthesis are delayed for several hours while mTOR-mediated events can be rapidly upregulated during this period (Nader & Esser, 2001). Eventual increases in protein synthesis appear to coincide with later eIF2B changes (Farrell et al. 1999). Without question, chronic mTOR signalling is indispensable for mediating increased cell size/muscle mass as inhibition of this pathway almost completely blocks the response (Bodine et al. 2001). Additionally, the downstream mTOR target, S6K1, is strongly linked with muscle hypertrophy (Baar & Esser, 1999). However, acute suppression of mTOR does not appear to dramatically affect overall rates of protein synthesis in skeletal muscle. Collectively, it is accurate to propose that both components of translation initiation are essential to increases in skeletal muscle mass. Events associated with eIF2B regulation may orchestrate the acute changes in protein synthesis following resistance exercise, whereas activation of mTOR/4E-BP1/S6K1 may result in preferential synthesis of proteins necessary to enhance the translational apparatus and optimize the capacity for protein synthesis with long-term training.
Rapid cell signalling and resistance exercise
Recent efforts to better understand regulation of translation initiation following an acute bout of resistance exercise suggest alterations wherein distinct eIF proteins are rapidly phosphorylated (Bolster et al. 2003). Intermittent and transient activation of these proteins may provide more precise control for modulating a growth response. Specifically, these responses appear temporal in nature and the acute impact of resistance exercise on mRNA translation likely becomes cumulative with each successive bout of exercise; the implication being this growth pathway is intermittently turned ‘on’ with repetitive resistance exercise and distinct mRNAs (ribosomal proteins, etc.) may accumulate to a point where an increase in the amount of specific proteins occurs (Neufer & Dohm, 1993). These responses highlight the longer-term and more rapid control mechanisms associated with transcription and translation, respectively that contribute to achieving muscle hypertrophy (Fig. 2).
Where do we go from here?
Identifying the key factors that rapidly initiate the cascade of signalling events in response to acute resistance exercise remain elusive but several candidates may include integrin activation and/or calcium mobilization. Integrins are transmembrane proteins that couple physical or chemical stimuli to intracellular events, whereas increased calcium flux is evident during muscle contraction. The rapid, yet transient upregulation of the pathway demonstrated by resistance exercise suggests these factors could quickly stimulate mTOR signalling which may then lead to select mRNA translation of local growth factors (i.e. IGF-1), further upregulating the growth response by the cell.
As we look to the future as to how our understanding of muscle hypertrophy will evolve it is important to acknowledge challenges as well. Comparison of data in this area of research is often difficult given the multitude of hypertrophy models currently employed. Tight correlation between cell culture and animal or human data does not always exist. Furthermore, the overlap of various signalling pathways and the rapidly expanding involvement of newly identified proteins makes interpretation complicated. The growth response by the cell incorporates multiple signalling inputs. An integrated response is therefore required and, however tempting, hypertrophy will likely never be isolated to one key protein acting as a ‘master switch’. Thus, a multi-faceted approach using genomic, proteomic and bioinformatic tools will be compulsory to elucidate the gaps in our knowledge of how muscle hypertrophy occurs.
References
Baar K & Esser KA (1999). Phosphorylation of p70S6k correlates with increased skeletal muscle mass following resistance exercise. Am J Physiol 276, C120-C127.
Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ & Yancopoulous GD (2001). Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nature Cell Biol 3, 1014-1019.
Bolster DR, Kubica N, Crozier SJ, Williamson DL, Farrell PA, Kimball SR & Jefferson LS (2003). Immediate response of mammalian target of rapamycin (mTOR)-mediated signalling following acute resistance exercise in rat skeletal muscle. J Physiol 553.1, 213-220.
Farrell PA, Fedele MJ, Vary TC, Kimball SR, Lang CH & Jefferson LS (1999). Regulation of protein synthesis after acute resistance exercise in diabetic rats. Am J Physiol 276, E721-E727.
Nader GA & Esser KA (2001). Intracellular signaling specificity in skeletal muscle in response to different modes of exercise. J Appl Physiol 90, 1936-1942.
Neufer PD & Dohm GL (1993). Exercise induces a transient increase in transcription of the GLUT-4 gene in skeletal muscle. Am J Physiol Cell Physiol 265, C1597-1603.
