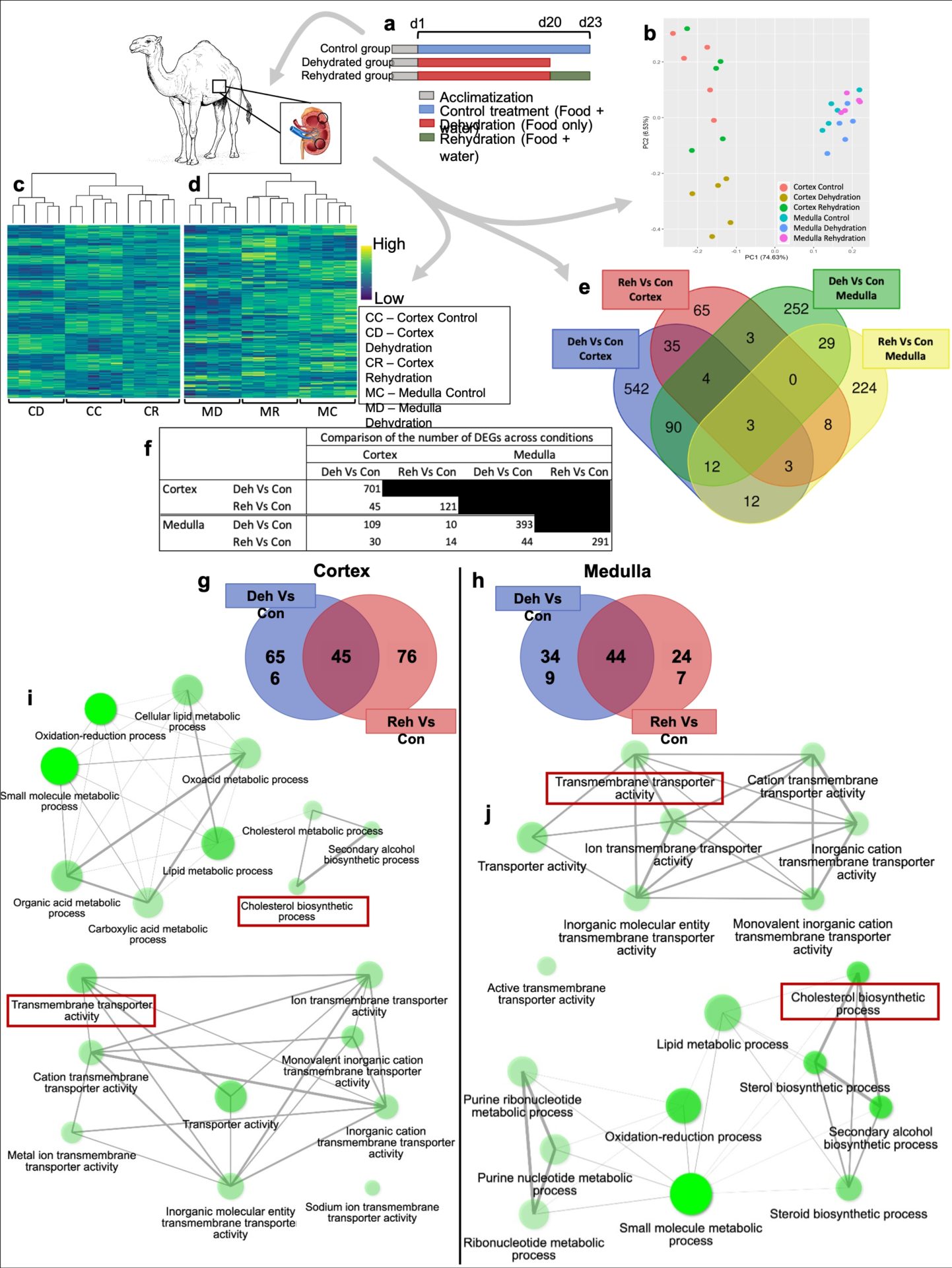
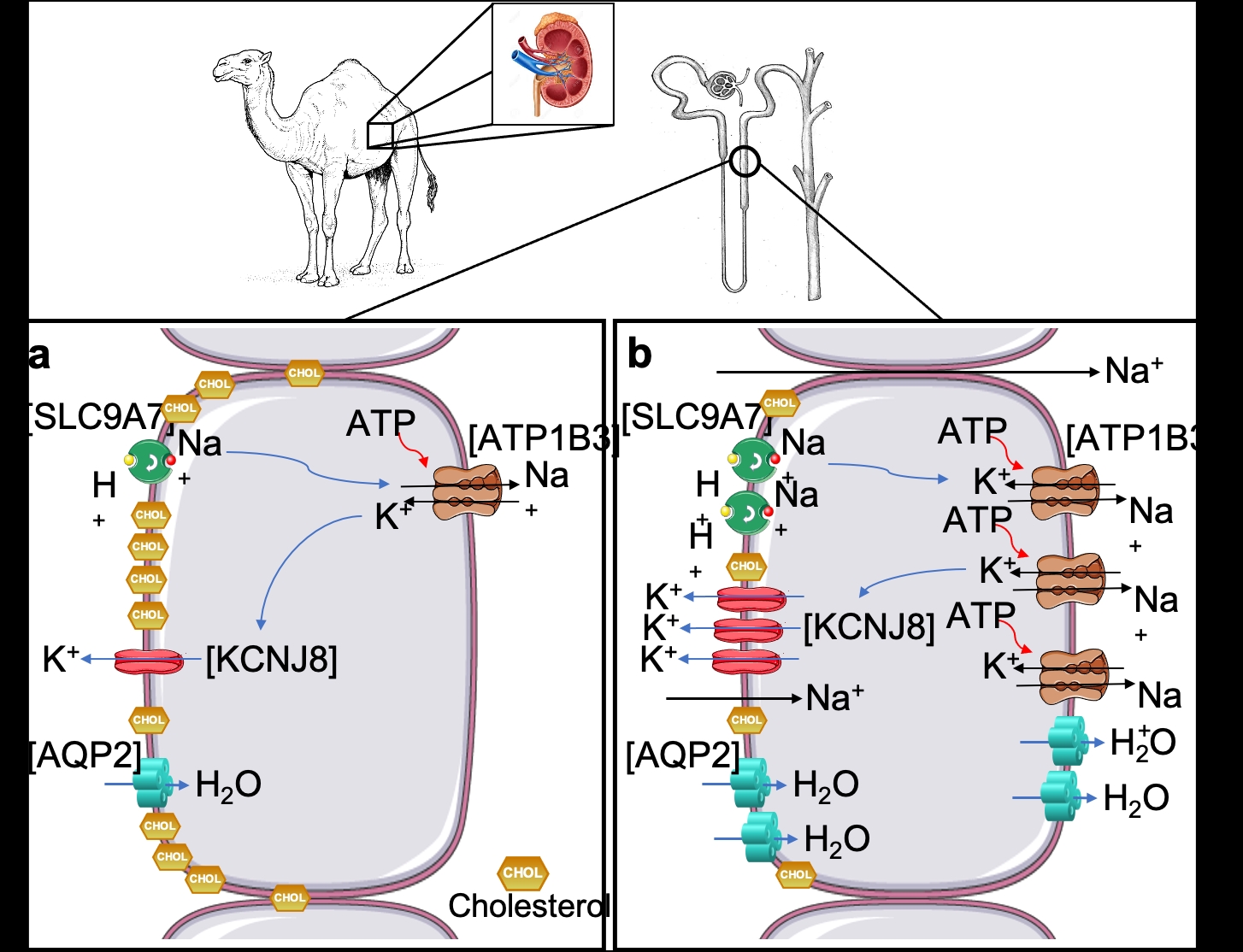
The one-humped Arabian camel is the most important livestock animal in arid and semi-arid regions and continues to provide basic necessities to millions of people in areas where food production is problematic. In the current context of global warming, there is renewed interest in the adaptive mechanisms that enable camelids to survive in arid conditions. Extensive evidence shows the impressive set of adaptations that allows dromedaries to thrive in such environment and recent investigations described genomic signatures that revealed evolutionary adaptations to desert environments (1-2). We now present a comprehensive catalogue of the transcriptomes of the male dromedary kidney. We performed RNAseq in samples from 15 adult dromedaries (5 controls, 5 dehydrated and 5 rehydrated). Using DESeq2, we identified 985 DEG in the kidney (cortex and medulla) of dehydrated camels. Gene Ontology (GO) analyses suggested an enrichment of the cholesterol biosynthetic pathway and an overrepresentation of categories related to “ion transmembrane transport”. The RNAseq data confirmed that 13 key genes involved in the cholesterol biosynthesis pathway were significantly (padj < 0.05) downregulated in the kidney medulla during dehydration. These data were validated by RT-qPCR (log2FC ranged 0.34 – 0.87). Further, we recently reconstructed the transcriptional regulatory network of the camel kidney during chronic dehydration. We identified 49 differentially expressed transcription factors and 10 enriched regulons. Among those, SREBF1 and SREBF2 were significantly downregulated in dehydration compared to control (padj < 0.05). They play a key role in the regulation of cholesterol synthesis and GO analyses of their significantly changed targets confirmed an enrichment of that pathway. Based on our hypothesis of a role for cholesterol during dehydration, we identified DEGs with known roles in the countercurrent multiplication process which are affected by changes in the level of cholesterol. Cholesterol changes the state of K+ inward rectifiers (KCNJ8), by directly binding onto it, to “silent”, it also inhibits Na+ leakage and indirectly decreases Na+-K+-ATPase activity. Moreover, cholesterol depletion results in the accumulation of AQP2 in the plasma membrane. KCNJ8 (padj < 0.01) and ATP1B3 (non-significant) showed a ~0.5-2 fold increase in gene expression during dehydration in the medulla, whilst SLC9A7 expression level trended towards a smaller increase (non-significant). Relative expression of AQP2 showed a 2-fold increase (padj < 0.001). Our datasets suggest that the suppression of cholesterol biosynthesis may facilitate water retention in the kidney of the dromedary by indirectly enhancing the osmotic gradient along the medullary interstitium that is required for the subsequent AQP-2-mediated water reabsorption. Ethical statement and statistics Samples were shipped to the University of Bristol under the auspices of a DEFRA Import Licence (TARP/2016/063). This study was approved by the Animal Ethics Committee of the United Arab Emirates University (approval ID: AE/15/38) and the University of Bristol Animal Welfare and Ethical Review Board. Benjamini-Hochberg was used to calculate significance levels in differential gene expression analysis and One-way ANOVA to determined differences between RT-qPCR experimental groups (n=5). Alternatively, we used Kruskal-Wallis test combined with Benjamini-Hochberg method for groups which followed non-normal distribution.
Future Physiology 2021 (Virutal) (2021) Proc Physiol Soc 47, OC21
Oral Communications: A comprehensive analysis of gene expression in the kidney of the one-humped Arabian camel (Camelus dromedarius) reveals a role for cholesterol in water conservation during dehydration
Benjamin T. Gillard1, Fernando Alvira-Iraizoz1, Panjiao Lin1, Alex Paterson1, Audrys Pauza1, Mahmoud Ali2, Ammar Hassib Alabsi3, Pamela Burger4, Naserddine Hamadi5, Abdu Adem2, David Murphy1, Michael Greenwood1
1 University of Bristol, Bristol, United Kingdom 2 United Arab Emirates University, Al-ain, The United Arab Emirates 3 Alfaisal University, Riyadh, Saudi Arabia 4 Vetmeduni, Vienna, Austria 5 College of Natural and Health Sciences, Abu Dhabi, The United Arab Emirates
View other abstracts by:
Where applicable, experiments conform with Society ethical requirements.