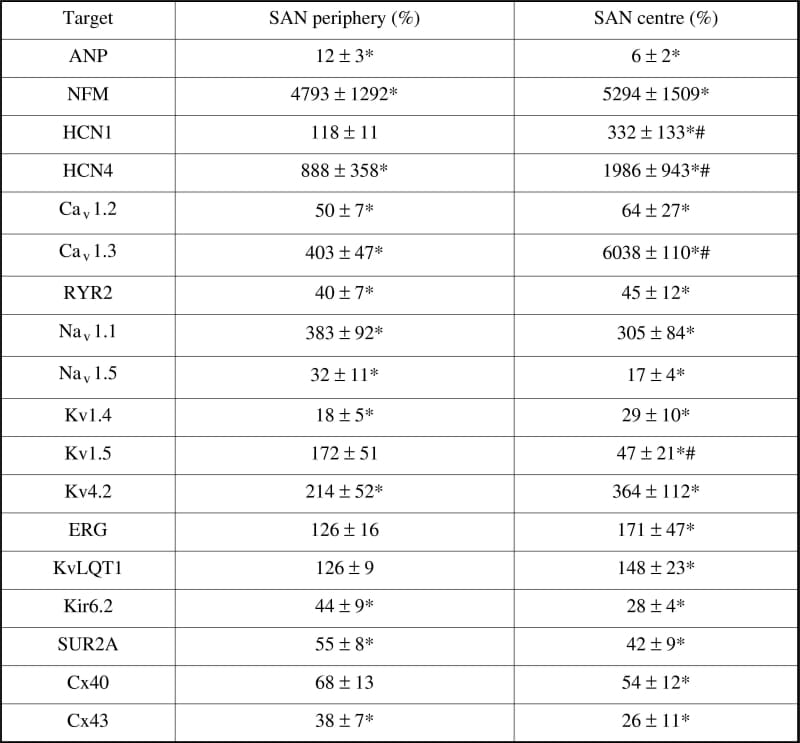
The sinoatrial node (SAN) is the pacemaker of the heart. Although the ionic currents responsible for pacemaker activity have been recorded from rabbit SAN, little is known about the molecular basis of these ionic currents. The aim of our study was to measure levels of mRNA coding for specific ion channels and their associated subunits in the rabbit SAN and surrounding atrial tissue. Rabbits were killed humanely and the SAN isolated. For quantitative PCR, cDNA was synthesised from tissue dissected from the centre and periphery of the SAN, as well as from the right atrium (RA; n=7 rabbits). All PCRs were performed in triplicate. Table 1 summarises the transcripts for which cDNA levels were significantly different between tissue samples. No significant differences in mRNA levels were found for Kv4.3, KChIP2, Kir2.1, Kir2.2, Kir3.1, Cx45, NCX1, SERCA2a, 28S and GAPDH. In situ hybridisation was performed with probes targeted to specific transcripts. The localisation of the in situ hybridisation labelling was in the main consistent with the quantitative PCR data. These results show that mRNA levels for ion channels and associated regulatory subunits vary within the rabbit SA node and between the SA node and surrounding atrial tissue. The pattern of expression of ion channels helps explain the ionic currents recorded.
University of Bristol (2005) J Physiol 567P, PC30
Poster Communications: Molecular mapping of the rabbit sinoatrial node
Tellez, James; Dobrzynski, H; Greener, I D; Graham, G M; Honjo, H; Yamamoto, M; Boyett, M R; Billeter, R;
1. University of Manchester, Manchester, United Kingdom. 2. University of Leeds, Leeds, United Kingdom. 3. Nagoya University, Nagoya, Japan.
View other abstracts by:
Table 1. Number of cDNA molecules relative to number of cDNA molecules in RA (100 %).*Significant difference compared to RA (P<0.05) #significant difference compared to SAN periphery (P<0.05). Significance of differences was calculated using one-way ANVOA. Data are shown as means ± S.E.M.
Where applicable, experiments conform with Society ethical requirements.