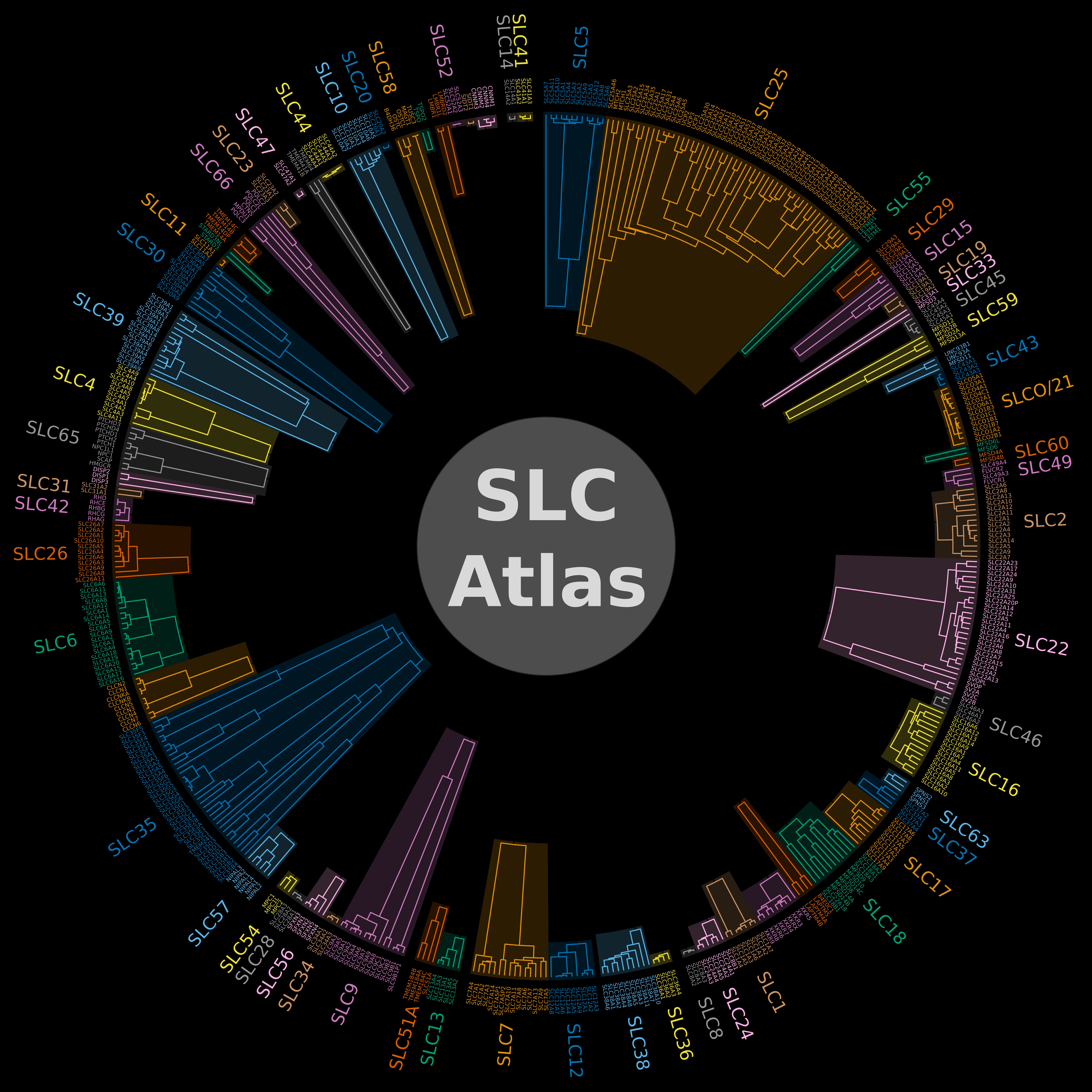
Background and aims: Transport of solutes across various biological membranes is essential to maintain cellular homeostasis and metabolism, and its dysfunction plays a pivotal role in the development of various diseases. The Solute Carrier (SLC) superfamily represents the largest and most diverse group of membrane transporter proteins, raising significant challenges in their identification, classification and annotation. Heterogeneity of SLC members also manifests at the level of protein structures, leading to several distinct structural folds among SLC transporters. Due to the absence of conserved sequence or structural signature motifs, we hypothesized that yet unidentified SLC transporters could exist in the human genome. In our work, we have undertaken a systematic meta-analysis of available data and literature in order to discover SLC-like proteins not yet in the official nomenclature. Contrary to similar analyses, we have strived to also find SLC-like proteins that are markedly dissimilar in sequence to the currently annotated ones, as well as use available structural information to define SLC superfamilies. A complete view of the human SLC-ome will play an instrumental role in understanding human physiology and can potentially be exploited for therapeutic benefits.
Methods: As a basis of our analysis, the Transporter Classification Database (TCDB), Protein families (Pfam), Uni-Prot, Protein Data Bank (PDB) databases have been used. Sequence similarity search was carried out using sequence profile hidden Markov-models (HMMs), using either models built by ourselves for individual TCDB protein families, or models obtained from Pfam.
Results: In order to perform a top-down search of SLC-like proteins, we have derived a set of eight criteria defining “SLC-likeness” in terms of properties that can be extracted from available databases. Manual curation of TCDB protein families and corresponding Pfam models was carried out based on the textual description of the families at the TCDB and Pfam web sites, respectively, in order to filter proteins and Pfam models that violate any of our SLC-likeness criteria. The remaining 166 protein families and 217 Pfam models were then used in sequence similarity searches against the proteomes of seven clinically relevant organisms, including human, rat, and mouse. The resulting 3669 proteins, including 520 from human, have subsequently been classified into families based on their pattern of similarity (fingerprint) to individual HMMs used in the search. Our analysis gave ~120 additional (“novel”), potentially SLC-like proteins compared to previously annotated SLCs, as well as ~40 additional protein families. Subsequent literature search on the found human proteins revealed that 53 of the “novel” SLC-like proteins could be assigned a small-molecule substrate.
Conclusions: The “newly” found transporters represent proteins that might have received less attention from the scientific community due to being missing from the official SLC nomenclature. In addition, several other putative SLC-like transporter proteins have been found. Subsequent analysis of structural homologs or predicted structures can identify further evolutionary relationships between the newly defined protein families. In summary, our results pave the way to a more unified view of the complete cellular “SLC-ome”, essential for a thorough understanding of fundamental physiological and pathological processes.